Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose
- PMID: 27702900
- PMCID: PMC5087047
- DOI: 10.1073/pnas.1608820113
Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose
Erratum in
-
Correction for Lever et al., Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose.Proc Natl Acad Sci U S A. 2017 Jan 10;114(2):E267. doi: 10.1073/pnas.1620047114. Epub 2017 Jan 3. Proc Natl Acad Sci U S A. 2017. PMID: 28049822 Free PMC article. No abstract available.
Abstract
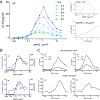
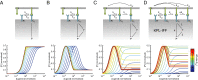
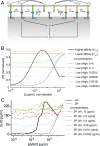
T cells must respond differently to antigens of varying affinity presented at different doses. Previous attempts to map peptide MHC (pMHC) affinity onto T-cell responses have produced inconsistent patterns of responses, preventing formulations of canonical models of T-cell signaling. Here, a systematic analysis of T-cell responses to 1 million-fold variations in both pMHC affinity and dose produced bell-shaped dose-response curves and different optimal pMHC affinities at different pMHC doses. Using sequential model rejection/identification algorithms, we identified a unique, minimal model of cellular signaling incorporating kinetic proofreading with limited signaling coupled to an incoherent feed-forward loop (KPL-IFF) that reproduces these observations. We show that the KPL-IFF model correctly predicts the T-cell response to antigen copresentation. Our work offers a general approach for studying cellular signaling that does not require full details of biochemical pathways.
Keywords: T-cell receptor; immunology; pathway architecture; signaling; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

