Ohgata, the Single Drosophila Ortholog of Human Cereblon, Regulates Insulin Signaling-dependent Organismic Growth
- PMID: 27702999
- PMCID: PMC5122779
- DOI: 10.1074/jbc.M116.757823
Ohgata, the Single Drosophila Ortholog of Human Cereblon, Regulates Insulin Signaling-dependent Organismic Growth
Abstract
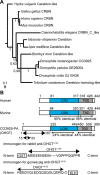
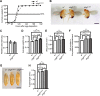
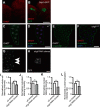
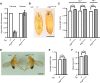
Cereblon (CRBN) is a substrate receptor of the E3 ubiquitin ligase complex that is highly conserved in animals and plants. CRBN proteins have been implicated in various biological processes such as development, metabolism, learning, and memory formation, and their impairment has been linked to autosomal recessive non-syndromic intellectual disability and cancer. Furthermore, human CRBN was identified as the primary target of thalidomide teratogenicity. Data on functional analysis of CRBN family members in vivo, however, are still scarce. Here we identify Ohgata (OHGT), the Drosophila ortholog of CRBN, as a regulator of insulin signaling-mediated growth. Using ohgt mutants that we generated by targeted mutagenesis, we show that its loss results in increased body weight and organ size without changes of the body proportions. We demonstrate that ohgt knockdown in the fat body, an organ analogous to mammalian liver and adipose tissue, phenocopies the growth phenotypes. We further show that overgrowth is due to an elevation of insulin signaling in ohgt mutants and to the down-regulation of inhibitory cofactors of circulating Drosophila insulin-like peptides (DILPs), named acid-labile subunit and imaginal morphogenesis protein-late 2. The two inhibitory proteins were previously shown to be components of a heterotrimeric complex with growth-promoting DILP2 and DILP5. Our study reveals OHGT as a novel regulator of insulin-dependent organismic growth in Drosophila.
Keywords: CRISPR/Cas; Cereblon; Drosophila genetics; E3 ubiquitin ligase; development; insulin.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

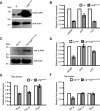
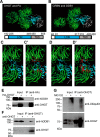
Similar articles
-
The E3 ubiquitin ligase component, Cereblon, is an evolutionarily conserved regulator of Wnt signaling.Nat Commun. 2021 Sep 6;12(1):5263. doi: 10.1038/s41467-021-25634-z. Nat Commun. 2021. PMID: 34489457 Free PMC article.
-
Cereblon in health and disease.Pflugers Arch. 2016 Aug;468(8):1299-309. doi: 10.1007/s00424-016-1854-1. Epub 2016 Jun 24. Pflugers Arch. 2016. PMID: 27343012 Review.
-
Ligand-mediated protein degradation reveals functional conservation among sequence variants of the CUL4-type E3 ligase substrate receptor cereblon.J Biol Chem. 2018 Apr 20;293(16):6187-6200. doi: 10.1074/jbc.M117.816868. Epub 2018 Feb 15. J Biol Chem. 2018. PMID: 29449372 Free PMC article.
-
Cereblon is recruited to aggresome and shows cytoprotective effect against ubiquitin-proteasome system dysfunction.Biochem Biophys Res Commun. 2015 Sep 4;464(4):1054-1059. doi: 10.1016/j.bbrc.2015.07.068. Epub 2015 Jul 15. Biochem Biophys Res Commun. 2015. PMID: 26188093
-
[Development of novel cereblon modulators and their target molecules].Rinsho Ketsueki. 2022;63(6):573-579. doi: 10.11406/rinketsu.63.573. Rinsho Ketsueki. 2022. PMID: 35831190 Review. Japanese.
Cited by
-
The E3 ubiquitin ligase component, Cereblon, is an evolutionarily conserved regulator of Wnt signaling.Nat Commun. 2021 Sep 6;12(1):5263. doi: 10.1038/s41467-021-25634-z. Nat Commun. 2021. PMID: 34489457 Free PMC article.
-
In vivo genome editing thrives with diversified CRISPR technologies.Zool Res. 2018 Mar 18;39(2):58-71. doi: 10.24272/j.issn.2095-8137.2017.012. Zool Res. 2018. PMID: 29515088 Free PMC article. Review.
-
Meep, a Novel Regulator of Insulin Signaling, Supports Development and Insulin Sensitivity via Maintenance of Protein Homeostasis in Drosophila melanogaster.G3 (Bethesda). 2020 Dec 3;10(12):4399-4410. doi: 10.1534/g3.120.401688. G3 (Bethesda). 2020. PMID: 32998936 Free PMC article.
-
The Neuroprotective Effect of Thalidomide against Ischemia through the Cereblon-mediated Repression of AMPK Activity.Sci Rep. 2018 Feb 6;8(1):2459. doi: 10.1038/s41598-018-20911-2. Sci Rep. 2018. PMID: 29410497 Free PMC article.
-
Pioneering protein degradation for agricultural applications.Commun Biol. 2025 Apr 9;8(1):591. doi: 10.1038/s42003-025-08013-y. Commun Biol. 2025. PMID: 40205025 Free PMC article.
References
-
- Garofalo R. S. (2002) Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 13, 156–162 - PubMed
-
- Edgar B. A. (2006) How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907–916 - PubMed
-
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., and Hafen E. (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 - PubMed
-
- Fuss B., Becker T., Zinke I., and Hoch M. (2006) The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature. 444, 945–948 - PubMed
-
- Ikeya T., Galic M., Belawat P., Nairz K., and Hafen E. (2002) Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293–1300 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

