Different DNA End Configurations Dictate Which NHEJ Components Are Most Important for Joining Efficiency
- PMID: 27703001
- PMCID: PMC5114395
- DOI: 10.1074/jbc.M116.752329
Different DNA End Configurations Dictate Which NHEJ Components Are Most Important for Joining Efficiency
Abstract
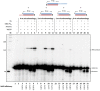
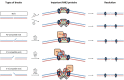
The nonhomologous DNA end-joining (NHEJ) pathway is a key mechanism for repairing dsDNA breaks that occur often in eukaryotic cells. In the simplest model, these breaks are first recognized by Ku, which then interacts with other NHEJ proteins to improve their affinity at DNA ends. These include DNA-PKcs and Artemis for trimming the DNA ends; DNA polymerase μ and λ to add nucleotides; and the DNA ligase IV complex to ligate the ends with the additional factors, XRCC4 (X-ray repair cross-complementing protein 4), XLF (XRCC4-like factor/Cernunos), and PAXX (paralog of XRCC4 and XLF). In vivo studies have demonstrated the degrees of importance of these NHEJ proteins in the mechanism of repair of dsDNA breaks, but interpretations can be confounded by other cellular processes. In vitro studies with NHEJ proteins have been performed to evaluate the nucleolytic resection, polymerization, and ligation steps, but a complete system has been elusive. Here we have developed a NHEJ reconstitution system that includes the nuclease, polymerase, and ligase components to evaluate relative NHEJ efficiency and analyze ligated junctional sequences for various types of DNA ends, including blunt, 5' overhangs, and 3' overhangs. We find that different dsDNA end structures have differential dependence on these enzymatic components. The dependence of some end joining on only Ku and XRCC4·DNA ligase IV allows us to formulate a physical model that incorporates nuclease and polymerase components as needed.
Keywords: DNA damage; DNA recombination; DNA repair; chromosomes; double-strand DNA break; enzyme; ligase; nuclease.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Martin G. M., Smith A. C., Ketterer D. J., Ogburn C. E., and Disteche C. M. (1985) Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Israel J. Med. Sci. 21, 296–301 - PubMed
-
- Rich T., Allen R. L., and Wyllie A. H. (2000) Defying death after DNA damage. Nature 407, 777–783 - PubMed
-
- Lieber M. R., and Karanjawala Z. E. (2004) Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell Biol. 5, 69–75 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

