Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma
- PMID: 27703150
- PMCID: PMC5059470
- DOI: 10.1038/ncomms12992
Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma
Erratum in
-
Erratum: Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma.Nat Commun. 2016 Nov 8;7:13591. doi: 10.1038/ncomms13591. Nat Commun. 2016. PMID: 27824035 Free PMC article. No abstract available.
Abstract
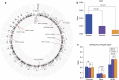
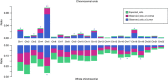
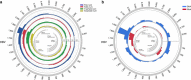
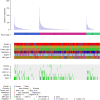
Hepatitis B virus (HBV) can integrate into the human genome, contributing to genomic instability and hepatocarcinogenesis. Here by conducting high-throughput viral integration detection and RNA sequencing, we identify 4,225 HBV integration events in tumour and adjacent non-tumour samples from 426 patients with HCC. We show that HBV is prone to integrate into rare fragile sites and functional genomic regions including CpG islands. We observe a distinct pattern in the preferential sites of HBV integration between tumour and non-tumour tissues. HBV insertional sites are significantly enriched in the proximity of telomeres in tumours. Recurrent HBV target genes are identified with few that overlap. The overall HBV integration frequency is much higher in tumour genomes of males than in females, with a significant enrichment of integration into chromosome 17. Furthermore, a cirrhosis-dependent HBV integration pattern is observed, affecting distinct targeted genes. Our data suggest that HBV integration has a high potential to drive oncogenic transformation.
Figures


References
-
- Forner A., Llovet J. M. & Bruix J. Hepatocellular carcinoma. Lancet 379, 1245–1255 (2012). - PubMed
-
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013). - PubMed
-
- Chen J. G. & Zhang S. W. Liver cancer epidemic in China: past, present and future. Semin. Cancer Biol. 21, 59–69 (2011). - PubMed
-
- Shafritz D. A. et al.. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. Studies in percutaneous liver biopsies and post-mortem tissue specimens. N. Engl. J. Med. 305, 1067–1073 (1981). - PubMed
-
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

