The SaeRS Two-Component System of Staphylococcus aureus
- PMID: 27706107
- PMCID: PMC5083920
- DOI: 10.3390/genes7100081
The SaeRS Two-Component System of Staphylococcus aureus
Abstract
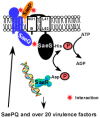
In the Gram-positive pathogenic bacterium Staphylococcus aureus, the SaeRS twocomponent system (TCS) plays a major role in controlling the production of over 20 virulence factors including hemolysins, leukocidins, superantigens, surface proteins, and proteases. The SaeRS TCS is composed of the sensor histidine kinase SaeS, response regulator SaeR, and two auxiliary proteins SaeP and SaeQ. Since its discovery in 1994, the sae locus has been studied extensively, and its contributions to staphylococcal virulence and pathogenesis have been well documented and understood; however, the molecular mechanism by which the SaeRS TCS receives and processes cognate signals is not. In this article, therefore, we review the literature focusing on the signaling mechanism and its interaction with other global regulators.
Keywords: Staphylococcus aureus; Bacterial histidine kinase; Two‐component system; Virulence factors.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, and in the decision to publish the results.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

