Mechanism of Rifampicin Inactivation in Nocardia farcinica
- PMID: 27706151
- PMCID: PMC5051949
- DOI: 10.1371/journal.pone.0162578
Mechanism of Rifampicin Inactivation in Nocardia farcinica
Abstract
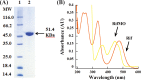
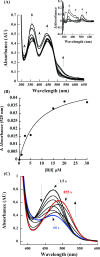
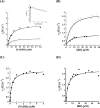
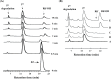
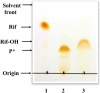
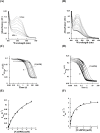
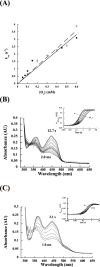
A novel mechanism of rifampicin (Rif) resistance has recently been reported in Nocardia farcinica. This new mechanism involves the activity of rifampicin monooxygenase (RifMO), a flavin-dependent monooxygenase that catalyzes the hydroxylation of Rif, which is the first step in the degradation pathway. Recombinant RifMO was overexpressed and purified for biochemical analysis. Kinetic characterization revealed that Rif binding is necessary for effective FAD reduction. RifMO exhibits only a 3-fold coenzyme preference for NADPH over NADH. RifMO catalyzes the incorporation of a single oxygen atom forming an unstable intermediate that eventually is converted to 2'-N-hydroxy-4-oxo-Rif. Stable C4a-hydroperoxyflavin was not detected by rapid kinetics methods, which is consistent with only 30% of the activated oxygen leading to product formation. These findings represent the first reported detailed biochemical characterization of a flavin-monooxygenase involved in antibiotic resistance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Lerner PI. Nocardiosis. Clin Infect Dis 1996:891–903. - PubMed
-
- McMurray DN. Mycobacteria and Nocardia. In: Baron S, editor. Medical Microbiology. 4th ed. Galveston (TX)1996. - PubMed
-
- Hartmann G, Honikel KO, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145(3):843–844. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

