Quantifying Heterogeneity in Host-Vector Contact: Tsetse (Glossina swynnertoni and G. pallidipes) Host Choice in Serengeti National Park, Tanzania
- PMID: 27706167
- PMCID: PMC5051720
- DOI: 10.1371/journal.pone.0161291
Quantifying Heterogeneity in Host-Vector Contact: Tsetse (Glossina swynnertoni and G. pallidipes) Host Choice in Serengeti National Park, Tanzania
Abstract
Background: Identifying hosts of blood-feeding insect vectors is crucial in understanding their role in disease transmission. Rhodesian human African trypanosomiasis (rHAT), also known as acute sleeping sickness is caused by Trypanosoma brucei rhodesiense and transmitted by tsetse flies. The disease is commonly associated with wilderness areas of east and southern Africa. Such areas hold a diverse range of species which form communities of hosts for disease maintenance. The relative importance of different wildlife hosts remains unclear. This study quantified tsetse feeding preferences in a wilderness area of great host species richness, Serengeti National Park, Tanzania, assessing tsetse feeding and host density contemporaneously.
Methods: Glossina swynnertoni and G. pallidipes were collected from six study sites. Bloodmeal sources were identified through matching Cytochrome B sequences amplified from bloodmeals from recently fed flies to published sequences. Densities of large mammal species in each site were quantified, and feeding indices calculated to assess the relative selection or avoidance of each host species by tsetse.
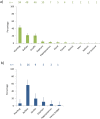
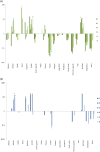
Results: The host species most commonly identified in G. swynnertoni bloodmeals, warthog (94/220), buffalo (48/220) and giraffe (46/220), were found at relatively low densities (3-11/km2) and fed on up to 15 times more frequently than expected by their relative density. Wildebeest, zebra, impala and Thomson's gazelle, found at the highest densities, were never identified in bloodmeals. Commonly identified hosts for G. pallidipes were buffalo (26/46), giraffe (9/46) and elephant (5/46).
Conclusions: This study is the first to quantify tsetse host range by molecular analysis of tsetse diet with simultaneous assessment of host density in a wilderness area. Although G. swynnertoni and G. pallidipes can feed on a range of species, they are highly selective. Many host species are rarely fed on, despite being present in areas where tsetse are abundant. These feeding patterns, along with the ability of key host species to maintain and transmit T. b. rhodesiense, drive the epidemiology of rHAT in wilderness areas.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Glossina dynamics in and around the sleeping sickness endemic Serengeti ecosystem of northwestern Tanzania.J Vector Ecol. 2007 Dec;32(2):263-8. doi: 10.3376/1081-1710(2007)32[263:gdiaat]2.0.co;2. J Vector Ecol. 2007. PMID: 18260516
-
Tracking the feeding patterns of tsetse flies (Glossina genus) by analysis of bloodmeals using mitochondrial cytochromes genes.PLoS One. 2011 Feb 28;6(2):e17284. doi: 10.1371/journal.pone.0017284. PLoS One. 2011. PMID: 21386971 Free PMC article.
-
Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in tsetse in Serengeti, Tanzania.PLoS Negl Trop Dis. 2012 Jan;6(1):e1501. doi: 10.1371/journal.pntd.0001501. Epub 2012 Jan 31. PLoS Negl Trop Dis. 2012. PMID: 22303496 Free PMC article.
-
Advancements in bait technology to control Glossina swynnertoni Austen, the species of limited distribution in Kenya and Tanzania border: A review.J Vector Borne Dis. 2017 Jan-Mar;54(1):16-24. J Vector Borne Dis. 2017. PMID: 28352042 Review.
-
Transmission Dynamics of Rhodesian Sleeping Sickness at the Interface of Wildlife and Livestock Areas.Trends Parasitol. 2016 Aug;32(8):608-621. doi: 10.1016/j.pt.2016.05.003. Epub 2016 Jun 1. Trends Parasitol. 2016. PMID: 27262917 Review.
Cited by
-
Parasites and blood-meal hosts of the tsetse fly in Tanzania: a metagenomics study.Parasit Vectors. 2022 Jun 22;15(1):224. doi: 10.1186/s13071-022-05344-1. Parasit Vectors. 2022. PMID: 35733222 Free PMC article.
-
Species richness and abundance of wild tsetse flies collected from selected human-wildlife-livestock interface in Tanzania.Parasite Epidemiol Control. 2024 Oct 25;27:e00389. doi: 10.1016/j.parepi.2024.e00389. eCollection 2024 Nov. Parasite Epidemiol Control. 2024. PMID: 39525368 Free PMC article.
-
Tsetse Bloodmeal Analyses Incriminate the Common Warthog Phacochoerus africanus as an Important Cryptic Host of Animal Trypanosomes in Smallholder Cattle Farming Communities in Shimba Hills, Kenya.Pathogens. 2021 Nov 18;10(11):1501. doi: 10.3390/pathogens10111501. Pathogens. 2021. PMID: 34832656 Free PMC article.
-
Tsetse blood-meal sources, endosymbionts and trypanosome-associations in the Maasai Mara National Reserve, a wildlife-human-livestock interface.PLoS Negl Trop Dis. 2021 Jan 6;15(1):e0008267. doi: 10.1371/journal.pntd.0008267. eCollection 2021 Jan. PLoS Negl Trop Dis. 2021. PMID: 33406097 Free PMC article.
-
Zebra skin odor repels the savannah tsetse fly, Glossina pallidipes (Diptera: Glossinidae).PLoS Negl Trop Dis. 2019 Jun 10;13(6):e0007460. doi: 10.1371/journal.pntd.0007460. eCollection 2019 Jun. PLoS Negl Trop Dis. 2019. PMID: 31181060 Free PMC article.
References
-
- Welburn SC, Picozzi K, Fevre EM, Coleman PG, Odiit M, et al. (2001) Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum- resistance-associated (SRA) gene. Lancet 358: 2017–2019. - PubMed
-
- Welburn SC, Picozzi K, Fyfe J, Fèvre E, Odiit M, et al. (2005) Control Options for Human Sleeping Sickness in Relation to the Animal Reservoir of Disease In: Osofsky SA, Cleaveland S, Karesh WB, Kock MD, Nyhus PJ, et al., editors. Conservation and Development Interventions at the Wildlife/Livestock Interface: Implications for Wildlife, Livestock and Human Health. IUCN, Gland, Switzerland and Cambridge, UK: pp. 55–61.
-
- Geigy R, Mwambu PM, Kauffman M (1971) Sleeping sickness survey in Musoma District, Tanzania: IV. Examination of wild mammals as a potential reservoir for T. rhodesiense. Acta Trop 28: 211–220. - PubMed
-
- Geigy R, Kauffman M (1973) Sleeping sickness survey in the Serengeti area (Tanzania) 1971: I. Examination of large mammals for trypanosomes. Acta Trop 30: 12–23. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

