Spatial and Temporal Resolution of Global Protein Synthesis during HSV Infection Using Bioorthogonal Precursors and Click Chemistry
- PMID: 27706239
- PMCID: PMC5051704
- DOI: 10.1371/journal.ppat.1005927
Spatial and Temporal Resolution of Global Protein Synthesis during HSV Infection Using Bioorthogonal Precursors and Click Chemistry
Abstract
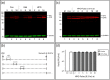
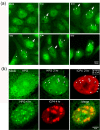
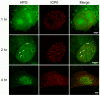
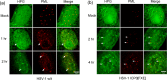
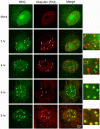
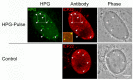
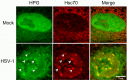
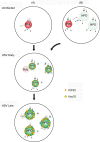
We used pulse-labeling with the methionine analogue homopropargylglycine (HPG) to investigate spatiotemporal aspects of protein synthesis during herpes simplex virus (HSV) infection. In vivo incorporation of HPG enables subsequent selective coupling of fluorochrome-capture reagents to newly synthesised proteins. We demonstrate that HPG labeling had no effect on cell viability, on accumulation of test early or late viral proteins, or on overall virus yields. HPG pulse-labeling followed by SDS-PAGE analysis confirmed incorporation into newly synthesised proteins, while parallel processing by in situ cycloaddition revealed new insight into spatiotemporal aspects of protein localisation during infection. A striking feature was the rapid accumulation of newly synthesised proteins not only in a general nuclear pattern but additionally in newly forming sub-compartments represented by small discrete foci. These newly synthesised protein domains (NPDs) were similar in size and morphology to PML domains but were more numerous, and whereas PML domains were progressively disrupted, NPDs were progressively induced and persisted. Immediate-early proteins ICP4 and ICP0 were excluded from NPDs, but using an ICP0 mutant defective in PML disruption, we show a clear spatial relationship between NPDs and PML domains with NPDs frequently forming immediately adjacent and co-joining persisting PML domains. Further analysis of location of the chaperone Hsc70 demonstrated that while NPDs formed early in infection without overt Hsc70 recruitment, later in infection Hsc70 showed pronounced recruitment frequently in a coat-like fashion around NPDs. Moreover, while ICP4 and ICP0 were excluded from NPDs, ICP22 showed selective recruitment. Our data indicate that NPDs represent early recruitment of host and viral de novo translated protein to distinct structural entities which are precursors to the previously described VICE domains involved in protein quality control in the nucleus, and reveal new features from which we propose spatially linked platforms of newly synthesised protein processing after nuclear import.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures







References
-
- Flint SJ, Enquist LW, Krug RM, Racaniello VR, Skalka AM (2009) Principles of Virology. Washington DC: ASM Press.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

