A Specially Designed Multi-Gene Panel Facilitates Genetic Diagnosis in Children with Intrahepatic Cholestasis: Simultaneous Test of Known Large Insertions/Deletions
- PMID: 27706244
- PMCID: PMC5051675
- DOI: 10.1371/journal.pone.0164058
A Specially Designed Multi-Gene Panel Facilitates Genetic Diagnosis in Children with Intrahepatic Cholestasis: Simultaneous Test of Known Large Insertions/Deletions
Abstract
Background and aims: Large indels are commonly identified in patients but are not detectable by routine Sanger sequencing and panel sequencing. We specially designed a multi-gene panel that could simultaneously test known large indels in addition to ordinary variants, and reported the diagnostic yield in patients with intrahepatic cholestasis.
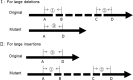
Methods: The panel contains 61 genes associated with cholestasis and 25 known recurrent large indels. The amplicon library was sequenced on Ion PGM system. Sequencing data were analyzed using a routine data analysis protocol and an internal program encoded for large indels test simultaneously. The validation phase was performed using 54 patients with known genetic diagnosis, including 5 with large insertions. At implement phase, 141 patients with intrahepatic cholestasis were evaluated.
Results: At validation phase, 99.6% of the variations identified by Sanger sequencing could be detected by panel sequencing. Following the routine protocol, 99.8% of false positives could be filtered and 98.8% of retained variations were true positives. Large insertions in the 5 patients with known genetic diagnosis could be correctly detected using the internal program. At implementation phase, 96.9% of the retained variations, following the routine protocol, were confirmed to be true. Twenty-nine patients received a potential genetic diagnosis when panel sequencing data were analyzed using the routine protocol. Two additional patients, who were found to harbor large insertions in SLC25A13, obtained a potential genetic diagnosis when sequencing data were further analyzed using the internal program. A total of 31 (22.0%) patients obtained a potential genetic diagnosis. Nine different genetic disorders were diagnosed, and citrin deficiency was the commonest.
Conclusion: Specially designed multi-gene panel can correctly detect large indels simultaneously. By using it, we assigned a potential genetic diagnosis to 22.0% of patients with intrahepatic cholestasis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Gordo-Gilart R, Andueza S, Hierro L, Martínez-Fernández P, D'Agostino D, Jara P, et al. Functional analysis of ABCB4 mutations relates clinical outcomes of progressive familial intrahepatic cholestasis type 3 to the degree of MDR3 floppase activity. Gut. 2015;64:147–55. 10.1136/gutjnl-2014-306896 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

