Heterodimers as the Structural Unit of the T=1 Capsid of the Fungal Double-Stranded RNA Rosellinia necatrix Quadrivirus 1
- PMID: 27707923
- PMCID: PMC5126378
- DOI: 10.1128/JVI.01013-16
Heterodimers as the Structural Unit of the T=1 Capsid of the Fungal Double-Stranded RNA Rosellinia necatrix Quadrivirus 1
Abstract
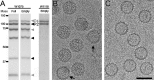
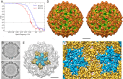
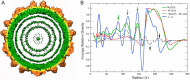
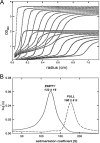
Most double-stranded RNA (dsRNA) viruses are transcribed and replicated in a specialized icosahedral capsid with a T=1 lattice consisting of 60 asymmetric capsid protein (CP) dimers. These capsids help to organize the viral genome and replicative complex(es). They also act as molecular sieves that isolate the virus genome from host defense mechanisms and allow the passage of nucleotides and viral transcripts. Rosellinia necatrix quadrivirus 1 (RnQV1), the type species of the family Quadriviridae, is a dsRNA fungal virus with a multipartite genome consisting of four monocistronic segments (segments 1 to 4). dsRNA-2 and dsRNA-4 encode two CPs (P2 and P4, respectively), which coassemble into ∼450-Å-diameter capsids. We used three-dimensional cryo-electron microscopy combined with complementary biophysical techniques to determine the structures of RnQV1 virion strains W1075 and W1118. RnQV1 has a quadripartite genome, and the capsid is based on a single-shelled T=1 lattice built of P2-P4 dimers. Whereas the RnQV1-W1118 capsid is built of full-length CP, P2 and P4 of RnQV1-W1075 are cleaved into several polypeptides, maintaining the capsid structural organization. RnQV1 heterodimers have a quaternary organization similar to that of homodimers of reoviruses and other dsRNA mycoviruses. The RnQV1 capsid is the first T=1 capsid with a heterodimer as an asymmetric unit reported to date and follows the architectural principle for dsRNA viruses that a 120-subunit capsid is a conserved assembly that supports dsRNA replication and organization.
Importance: Given their importance to health, members of the family Reoviridae are the basis of most structural and functional studies and provide much of our knowledge of dsRNA viruses. Analysis of bacterial, protozoal, and fungal dsRNA viruses has improved our understanding of their structure, function, and evolution, as well. Here, we studied a dsRNA virus that infects the fungus Rosellinia necatrix, an ascomycete that is pathogenic to a wide range of plants. Using three-dimensional cryo-electron microscopy and analytical ultracentrifugation analysis, we determined the structure and stoichiometry of Rosellinia necatrix quadrivirus 1 (RnQV1). The RnQV1 capsid is a T=1 capsid with 60 heterodimers as the asymmetric units. The large amount of genetic information used by RnQV1 to construct a simple T=1 capsid is probably related to the numerous virus-host and virus-virus interactions that it must face in its life cycle, which lacks an extracellular phase.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.PLoS Pathog. 2017 Dec 8;13(12):e1006755. doi: 10.1371/journal.ppat.1006755. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29220409 Free PMC article.
-
Capsid Structure of dsRNA Fungal Viruses.Viruses. 2018 Sep 7;10(9):481. doi: 10.3390/v10090481. Viruses. 2018. PMID: 30205532 Free PMC article. Review.
-
Structure and assembly of double-stranded RNA mycoviruses.Adv Virus Res. 2020;108:213-247. doi: 10.1016/bs.aivir.2020.08.001. Epub 2020 Sep 16. Adv Virus Res. 2020. PMID: 33837717
-
Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.J Virol. 2020 Nov 23;94(24):e01542-20. doi: 10.1128/JVI.01542-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32938763 Free PMC article.
-
Chrysovirus structure: repeated helical core as evidence of gene duplication.Adv Virus Res. 2013;86:87-108. doi: 10.1016/B978-0-12-394315-6.00004-0. Adv Virus Res. 2013. PMID: 23498904 Review.
Cited by
-
Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.mBio. 2021 Mar 30;12(2):e02924-20. doi: 10.1128/mBio.02924-20. mBio. 2021. PMID: 33785622 Free PMC article.
-
Identification, Molecular Characterization, and Biology of a Novel Quadrivirus Infecting the Phytopathogenic Fungus Leptosphaeria biglobosa.Viruses. 2018 Dec 25;11(1):9. doi: 10.3390/v11010009. Viruses. 2018. PMID: 30585188 Free PMC article.
-
IRAM: virus capsid database and analysis resource.Database (Oxford). 2019 Jan 1;2019:baz079. doi: 10.1093/database/baz079. Database (Oxford). 2019. PMID: 31318422 Free PMC article.
-
Phosphatidylinositol 3-Phosphate Mediates the Establishment of Infectious Bursal Disease Virus Replication Complexes in Association with Early Endosomes.J Virol. 2021 Feb 24;95(6):e02313-20. doi: 10.1128/JVI.02313-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361427 Free PMC article.
-
Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.PLoS Pathog. 2017 Dec 8;13(12):e1006755. doi: 10.1371/journal.ppat.1006755. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29220409 Free PMC article.
References
-
- Patton JT. 2008. Segmented double-stranded RNA viruses: structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
-
- Steven AC, Baumeister W, Johnson LN, OPerham RN. 2016. Molecular biology of assemblies and machines. Garland Science, New York, NY.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

