Repression of the Chromatin-Tethering Domain of Murine Leukemia Virus p12
- PMID: 27707926
- PMCID: PMC5126376
- DOI: 10.1128/JVI.01084-16
Repression of the Chromatin-Tethering Domain of Murine Leukemia Virus p12
Abstract
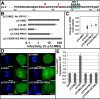
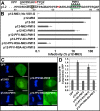
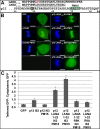
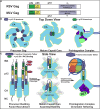
Murine leukemia virus (MLV) p12, encoded within Gag, binds the viral preintegration complex (PIC) to the mitotic chromatin. This acts to anchor the viral PIC in the nucleus as the nuclear envelope re-forms postmitosis. Mutations within the p12 C terminus (p12 PM13 to PM15) block early stages in viral replication. Within the p12 PM13 region (p12 60PSPMA65), our studies indicated that chromatin tethering was not detected when the wild-type (WT) p12 protein (M63) was expressed as a green fluorescent protein (GFP) fusion; however, constructs bearing p12-I63 were tethered. N-terminal truncations of the activated p12-I63-GFP indicated that tethering increased further upon deletion of p12 25DLLTEDPPPY34, which includes the late domain required for viral assembly. The p12 PM15 sequence (p12 70RREPP74) is critical for wild-type viral viability; however, virions bearing the PM15 mutation (p12 70AAAAA74) with a second M63I mutant were viable, with a titer 18-fold lower than that of the WT. The p12 M63I mutation amplified chromatin tethering and compensated for the loss of chromatin binding of p12 PM15. Rescue of the p12-M63-PM15 nonviable mutant with prototype foamy virus (PFV) and Kaposi's sarcoma herpesvirus (KSHV) tethering sequences confirmed the function of p1270-74 in chromatin binding. Minimally, full-strength tethering was seen with only p12 61SPIASRLRGRR71 fused to GFP. These results indicate that the p12 C terminus alone is sufficient for chromatin binding and that the presence of the p12 25DLLTEDPPPY34 motif in the N terminus suppresses the ability to tether.
Importance: This study defines a regulatory mechanism controlling the differential roles of the MLV p12 protein in early and late replication. During viral assembly and egress, the late domain within the p12 N terminus functions to bind host vesicle release factors. During viral entry, the C terminus of p12 is required for tethering to host mitotic chromosomes. Our studies indicate that the p12 domain including the PPPY late sequence temporally represses the p12 chromatin tethering motif. Maximal p12 tethering was identified with only an 11-amino-acid minimal chromatin tethering motif encoded at p1261-71 Within this region, the p12-M63I substitution switches p12 into a tethering-competent state, partially rescuing the p12-PM15 tethering mutant. A model for how this conformational change regulates early versus late functions is presented.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Phosphorylation Requirement of Murine Leukemia Virus p12.J Virol. 2016 Nov 28;90(24):11208-11219. doi: 10.1128/JVI.01178-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707931 Free PMC article.
-
Murine leukemia virus p12 tethers the capsid-containing pre-integration complex to chromatin by binding directly to host nucleosomes in mitosis.PLoS Pathog. 2018 Jun 15;14(6):e1007117. doi: 10.1371/journal.ppat.1007117. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29906285 Free PMC article.
-
Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses.J Virol. 2000 Aug;74(16):7250-60. doi: 10.1128/jvi.74.16.7250-7260.2000. J Virol. 2000. PMID: 10906179 Free PMC article.
-
Moloney Murine Leukemia Virus p12 Is Required for Histone Loading onto Retroviral DNAs.J Virol. 2021 Jul 12;95(15):e0049521. doi: 10.1128/JVI.00495-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011543 Free PMC article.
-
Early events in foamy virus-host interaction and intracellular trafficking.Viruses. 2013 Apr 8;5(4):1055-74. doi: 10.3390/v5041055. Viruses. 2013. PMID: 23567621 Free PMC article. Review.
Cited by
-
Targeting the Nucleosome Acidic Patch by Viral Proteins: Two Birds with One Stone?mBio. 2022 Apr 26;13(2):e0173321. doi: 10.1128/mbio.01733-21. Epub 2022 Mar 28. mBio. 2022. PMID: 35343785 Free PMC article. Review.
-
Evidence for Tethering of Human Cytomegalovirus Genomes to Host Chromosomes.Front Cell Infect Microbiol. 2020 Sep 30;10:577428. doi: 10.3389/fcimb.2020.577428. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33117732 Free PMC article.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. PMID: 38293010 Free PMC article. Updated. Preprint.
-
A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.Structure. 2021 Aug 5;29(8):886-898.e6. doi: 10.1016/j.str.2021.01.010. Epub 2021 Feb 15. Structure. 2021. PMID: 33592170 Free PMC article.
-
Phosphorylation Requirement of Murine Leukemia Virus p12.J Virol. 2016 Nov 28;90(24):11208-11219. doi: 10.1128/JVI.01178-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707931 Free PMC article.
References
-
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 110:12036–12041. doi:10.1073/pnas.1307157110. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

