Comparative Genomic Analysis of Haemophilus haemolyticus and Nontypeable Haemophilus influenzae and a New Testing Scheme for Their Discrimination
- PMID: 27707939
- PMCID: PMC5121393
- DOI: 10.1128/JCM.01511-16
Comparative Genomic Analysis of Haemophilus haemolyticus and Nontypeable Haemophilus influenzae and a New Testing Scheme for Their Discrimination
Abstract
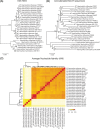
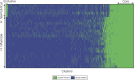
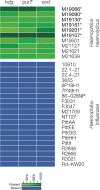
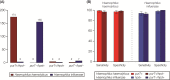
Haemophilus haemolyticus has been recently discovered to have the potential to cause invasive disease. It is closely related to nontypeable Haemophilus influenzae (NT H. influenzae). NT H. influenzae and H. haemolyticus are often misidentified because none of the existing tests targeting the known phenotypes of H. haemolyticus are able to specifically identify H. haemolyticus Through comparative genomic analysis of H. haemolyticus and NT H. influenzae, we identified genes unique to H. haemolyticus that can be used as targets for the identification of H. haemolyticus A real-time PCR targeting purT (encoding phosphoribosylglycinamide formyltransferase 2 in the purine synthesis pathway) was developed and evaluated. The lower limit of detection was 40 genomes/PCR; the sensitivity and specificity in detecting H. haemolyticus were 98.9% and 97%, respectively. To improve the discrimination of H. haemolyticus and NT H. influenzae, a testing scheme combining two targets (H. haemolyticus purT and H. influenzae hpd, encoding protein D lipoprotein) was also evaluated and showed 96.7% sensitivity and 98.2% specificity for the identification of H. haemolyticus and 92.8% sensitivity and 100% specificity for the identification of H. influenzae, respectively. The dual-target testing scheme can be used for the diagnosis and surveillance of infection and disease caused by H. haemolyticus and NT H. influenzae.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Simultaneous identification of Haemophilus influenzae and Haemophilus haemolyticus using real-time PCR.Future Microbiol. 2017 Jun;12:585-593. doi: 10.2217/fmb-2016-0215. Epub 2017 Jun 12. Future Microbiol. 2017. PMID: 28604066
-
Development of a diagnostic real-time polymerase chain reaction assay for the detection of invasive Haemophilus influenzae in clinical samples.Diagn Microbiol Infect Dis. 2012 Dec;74(4):356-62. doi: 10.1016/j.diagmicrobio.2012.08.018. Epub 2012 Sep 25. Diagn Microbiol Infect Dis. 2012. PMID: 23017260
-
Haemophilus haemolyticus is infrequently misidentified as Haemophilus influenzae in diagnostic specimens in Australia.Diagn Microbiol Infect Dis. 2014 Dec;80(4):272-3. doi: 10.1016/j.diagmicrobio.2014.08.016. Epub 2014 Sep 6. Diagn Microbiol Infect Dis. 2014. PMID: 25266675
-
Nontypeable Haemophilus influenzae as a pathogen in children.Pediatr Infect Dis J. 2009 Jan;28(1):43-8. doi: 10.1097/INF.0b013e318184dba2. Pediatr Infect Dis J. 2009. PMID: 19057458 Review.
-
Invasive polyarticular septic arthritis caused by nontypeable haemophilus influenzae in a young adult: a case report and literature review.J Clin Rheumatol. 2011 Oct;17(7):380-2. doi: 10.1097/RHU.0b013e318236e499. J Clin Rheumatol. 2011. PMID: 21946466 Review.
Cited by
-
Draft Genome Sequences for a Diverse Set of Seven Haemophilus and Aggregatibacter Species.Microbiol Resour Announc. 2018 Oct 25;7(16):e00880-18. doi: 10.1128/MRA.00880-18. eCollection 2018 Oct. Microbiol Resour Announc. 2018. PMID: 30533737 Free PMC article.
-
Whole-Genome Sequencing of Aggregatibacter Species Isolated from Human Clinical Specimens and Description of Aggregatibacter kilianii sp. nov.J Clin Microbiol. 2018 Jun 25;56(7):e00053-18. doi: 10.1128/JCM.00053-18. Print 2018 Jul. J Clin Microbiol. 2018. PMID: 29695522 Free PMC article.
-
Comparative Genomic Analysis Reveals Genetic Diversity and Pathogenic Potential of Haemophilus seminalis and Emended Description of Haemophilus seminalis.Microbiol Spectr. 2023 Aug 17;11(4):e0477222. doi: 10.1128/spectrum.04772-22. Epub 2023 Jun 29. Microbiol Spectr. 2023. PMID: 37382545 Free PMC article.
-
Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease.Pathog Dis. 2017 Jun 1;75(4):ftx042. doi: 10.1093/femspd/ftx042. Pathog Dis. 2017. PMID: 28449098 Free PMC article. Review.
-
Patients with Chronic Obstructive Pulmonary Disease harbour a variation of Haemophilus species.Sci Rep. 2018 Oct 3;8(1):14734. doi: 10.1038/s41598-018-32973-3. Sci Rep. 2018. PMID: 30282975 Free PMC article. Clinical Trial.
References
-
- MacNeil JR, Cohn AC, Farley M, Mair R, Baumbach J, Bennett N, Gershman K, Harrison LH, Lynfield R, Petit S, Reingold A, Schaffner W, Thomas A, Coronado F, Zell ER, Mayer LW, Clark TA, Messonnier NE. 2011. Current epidemiology and trends in invasive Haemophilus influenzae disease—United States, 1989-2008. Clin Infect Dis 53:1230–1236. doi:10.1093/cid/cir735. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

