Exposure to the Functional Bacterial Amyloid Protein Curli Enhances Alpha-Synuclein Aggregation in Aged Fischer 344 Rats and Caenorhabditis elegans
- PMID: 27708338
- PMCID: PMC5052651
- DOI: 10.1038/srep34477
Exposure to the Functional Bacterial Amyloid Protein Curli Enhances Alpha-Synuclein Aggregation in Aged Fischer 344 Rats and Caenorhabditis elegans
Abstract
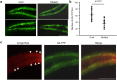
Misfolded alpha-synuclein (AS) and other neurodegenerative disorder proteins display prion-like transmission of protein aggregation. Factors responsible for the initiation of AS aggregation are unknown. To evaluate the role of amyloid proteins made by the microbiota we exposed aged rats and transgenic C. elegans to E. coli producing the extracellular bacterial amyloid protein curli. Rats exposed to curli-producing bacteria displayed increased neuronal AS deposition in both gut and brain and enhanced microgliosis and astrogliosis compared to rats exposed to either mutant bacteria unable to synthesize curli, or to vehicle alone. Animals exposed to curli producing bacteria also had more expression of TLR2, IL-6 and TNF in the brain than the other two groups. There were no differences among the rat groups in survival, body weight, inflammation in the mouth, retina, kidneys or gut epithelia, and circulating cytokine levels. AS-expressing C. elegans fed on curli-producing bacteria also had enhanced AS aggregation. These results suggest that bacterial amyloid functions as a trigger to initiate AS aggregation through cross-seeding and also primes responses of the innate immune system.
Figures


References
-
- Friedland R. P. Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. Journal of Alzheimer’s disease: JAD 45, 349–362 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

