Single cell dynamic phenotyping
- PMID: 27708391
- PMCID: PMC5052535
- DOI: 10.1038/srep34785
Single cell dynamic phenotyping
Abstract
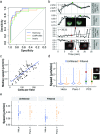
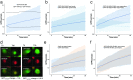
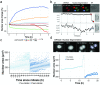
Live cell imaging has improved our ability to measure phenotypic heterogeneity. However, bottlenecks in imaging and image processing often make it difficult to differentiate interesting biological behavior from technical artifact. Thus there is a need for new methods that improve data quality without sacrificing throughput. Here we present a 3-step workflow to improve dynamic phenotype measurements of heterogeneous cell populations. We provide guidelines for image acquisition, phenotype tracking, and data filtering to remove erroneous cell tracks using the novel Tracking Aberration Measure (TrAM). Our workflow is broadly applicable across imaging platforms and analysis software. By applying this workflow to cancer cell assays, we reduced aberrant cell track prevalence from 17% to 2%. The cost of this improvement was removing 15% of the well-tracked cells. This enabled detection of significant motility differences between cell lines. Similarly, we avoided detecting a false change in translocation kinetics by eliminating the true cause: varied proportions of unresponsive cells. Finally, by systematically seeking heterogeneous behaviors, we detected subpopulations that otherwise could have been missed, including early apoptotic events and pre-mitotic cells. We provide optimized protocols for specific applications and step-by-step guidelines for adapting them to a variety of biological systems.
Figures

References
-
- Snijder B. & Pelkmans L. Origins of regulated cell-to-cell variability. Nat. Rev. Mol. Cell Biol. 12, 119–125 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

