Acetylation Mimics Within a Single Nucleosome Alter Local DNA Accessibility In Compacted Nucleosome Arrays
- PMID: 27708426
- PMCID: PMC5052607
- DOI: 10.1038/srep34808
Acetylation Mimics Within a Single Nucleosome Alter Local DNA Accessibility In Compacted Nucleosome Arrays
Abstract
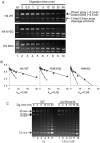
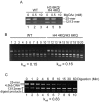
The activation of a silent gene locus is thought to involve pioneering transcription factors that initiate changes in the local chromatin structure to increase promoter accessibility and binding of downstream effectors. To better understand the molecular requirements for the first steps of locus activation, we investigated whether acetylation of a single nucleosome is sufficient to alter DNA accessibility within a condensed 25-nucleosome array. We found that acetylation mimics within the histone H4 tail domain increased accessibility of the surrounding linker DNA, with the increased accessibility localized to the immediate vicinity of the modified nucleosome. In contrast, acetylation mimics within the H3 tail had little effect, but were able to synergize with H4 tail acetylation mimics to further increase accessibility. Moreover, replacement of the central nucleosome with a nucleosome free region also resulted in increased local, but not global DNA accessibility. Our results indicate that modification or disruption of only a single target nucleosome results in significant changes in local chromatin architecture and suggest that very localized chromatin modifications imparted by pioneer transcription factors are sufficient to initiate a cascade of events leading to promoter activation.
Figures


References
-
- van Holde K. E. Chromatin (Springer: Verlag, , 1989).
-
- Tremethick D. J. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell 128, 651–654 (2007). - PubMed
-
- Hansen J. C. Conformational Dynamics of the Chromatin Fiber in Solution: Determinants, Mechanisms, and Functions. Annu Rev Biophys Biomol Struct 31, 361–392 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

