PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy
- PMID: 27708722
- PMCID: PMC5037587
- DOI: 10.1186/s13148-016-0270-x
PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy
Abstract
Background: Molecular biomarkers that might help to distinguish between more aggressive and clinically insignificant prostate cancers (PCa) are still urgently needed. Aberrant DNA methylation as a common molecular alteration in PCa seems to be a promising source for such biomarkers. In this study, PITX3 DNA methylation (mPITX3) and its potential role as a prognostic biomarker were investigated. Furthermore, mPITX3 was analyzed in combination with the established PCa methylation biomarker PITX2 (mPITX2).
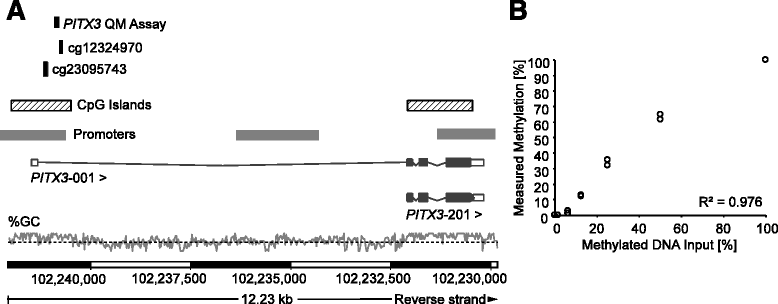
Methods: mPITX3 and mPITX2 were assessed by a quantitative real-time PCR and by means of the Infinium HumanMethylation450 BeadChip. BeadChip data were obtained from The Cancer Genome Atlas (TCGA) Research Network. DNA methylation differences between normal adjacent, benign hyperplastic, and carcinomatous prostate tissues were examined in the TCGA dataset as well as in prostatectomy specimens from the University Hospital Bonn. Retrospective analyses of biochemical recurrence (BCR) were conducted in a training cohort (n = 498) from the TCGA and an independent validation cohort (n = 300) from the University Hospital Bonn. All patients received radical prostatectomy.
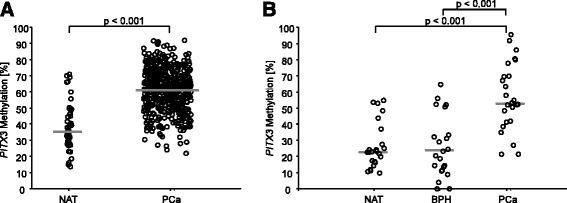
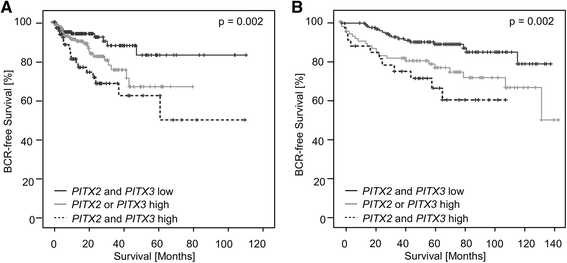
Results: In PCa tissue, mPITX3 was increased significantly compared to normal and benign hyperplastic tissue. In univariate Cox proportional hazards analyses, mPITX3 showed a significant prognostic value for BCR (training cohort: hazard ratio (HR) = 1.83 (95 % CI 1.07-3.11), p = 0.027; validation cohort: HR = 2.56 (95 % CI 1.44-4.54), p = 0.001). A combined evaluation with PITX2 methylation further revealed that hypermethylation of a single PITX gene member (either PITX2 or PITX3) identifies an intermediate risk group.
Conclusions: PITX3 DNA methylation alone and in combination with PITX2 is a promising biomarker for the risk stratification of PCa patients and adds relevant prognostic information to common clinically implemented parameters. Further studies are required to determine whether the results are transferable to a biopsy-based patient cohort. Trial registration: Patients for this unregistered study were enrolled retrospectively.
Keywords: DNA methylation; PITX2; PITX3; Prognostic biomarker; Prostate cancer.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

