The Role of Vitamin D in the Transcriptional Program of Human Pregnancy
- PMID: 27711190
- PMCID: PMC5053446
- DOI: 10.1371/journal.pone.0163832
The Role of Vitamin D in the Transcriptional Program of Human Pregnancy
Abstract
Background: Patterns of gene expression of human pregnancy are poorly understood. In a trial of vitamin D supplementation in pregnant women, peripheral blood transcriptomes were measured longitudinally on 30 women and used to characterize gene co-expression networks.
Objective: Studies suggest that increased maternal Vitamin D levels may reduce the risk of asthma in early life, yet the underlying mechanisms have not been examined. In this study, we used a network-based approach to examine changes in gene expression profiles during the course of normal pregnancy and evaluated their association with maternal Vitamin D levels.
Design: The VDAART study is a randomized clinical trial of vitamin D supplementation in pregnancy for reduction of pediatric asthma risk. The trial enrolled 881 women at 10-18 weeks of gestation. Longitudinal gene expression measures were obtained on thirty pregnant women, using RNA isolated from peripheral blood samples obtained in the first and third trimesters. Differentially expressed genes were identified using significance of analysis of microarrays (SAM), and clustered using a weighted gene co-expression network analysis (WGCNA). Gene-set enrichment was performed to identify major biological pathways.
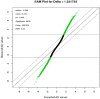
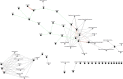
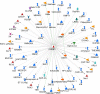
Results: Comparison of transcriptional profiles between first and third trimesters of pregnancy identified 5839 significantly differentially expressed genes (FDR<0.05). Weighted gene co-expression network analysis clustered these transcripts into 14 co-expression modules of which two showed significant correlation with maternal vitamin D levels. Pathway analysis of these two modules revealed genes enriched in immune defense pathways and extracellular matrix reorganization as well as genes enriched in notch signaling and transcription factor networks.
Conclusion: Our data show that gene expression profiles of healthy pregnant women change during the course of pregnancy and suggest that maternal Vitamin D levels influence transcriptional profiles. These alterations of the maternal transcriptome may contribute to fetal immune imprinting and reduce allergic sensitization in early life.
Trial registration: clinicaltrials.gov NCT00920621.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Montefort S, Ellul P, Montefort M, Caruana S, Grech V, Agius Muscat H. The effect of cigarette smoking on allergic conditions in Maltese children (ISAAC). Pediatric allergy and immunology: official publication of the European Society of Pediatric Allergy and Immunology. 2012;23(5):472–8. Epub 2012/03/23. 10.1111/j.1399-3038.2012.01276.x . - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

