A Comparison of the Costs and Benefits of Bacterial Gene Expression
- PMID: 27711251
- PMCID: PMC5053530
- DOI: 10.1371/journal.pone.0164314
A Comparison of the Costs and Benefits of Bacterial Gene Expression
Abstract
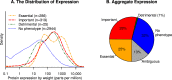
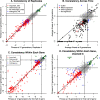
To study how a bacterium allocates its resources, we compared the costs and benefits of most (86%) of the proteins in Escherichia coli K-12 during growth in minimal glucose medium. The cost or investment in each protein was estimated from ribosomal profiling data, and the benefit of each protein was measured by assaying a library of transposon mutants. We found that proteins that are important for fitness are usually highly expressed, and 95% of these proteins are expressed at above 13 parts per million (ppm). Conversely, proteins that do not measurably benefit the host (with a benefit of less than 5% per generation) tend to be weakly expressed, with a median expression of 13 ppm. In aggregate, genes with no detectable benefit account for 31% of protein production, or about 22% if we correct for genetic redundancy. Although some of the apparently unnecessary expression could have subtle benefits in minimal glucose medium, the majority of the burden is due to genes that are important in other conditions. We propose that at least 13% of the cell's protein is "on standby" in case conditions change.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Discovery of glpC, an organic solvent tolerance-related gene in Escherichia coli, using gene expression profiles from DNA microarrays.Appl Environ Microbiol. 2005 Feb;71(2):1093-6. doi: 10.1128/AEM.71.2.1093-1096.2005. Appl Environ Microbiol. 2005. PMID: 15691972 Free PMC article.
-
A comparative analysis of industrial Escherichia coli K-12 and B strains in high-glucose batch cultivations on process-, transcriptome- and proteome level.PLoS One. 2013 Aug 8;8(8):e70516. doi: 10.1371/journal.pone.0070516. eCollection 2013. PLoS One. 2013. PMID: 23950949 Free PMC article.
-
Effect of simulated microgravity on E. coli K12 MG1655 growth and gene expression.PLoS One. 2013;8(3):e57860. doi: 10.1371/journal.pone.0057860. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472115 Free PMC article.
-
YeeI, a novel protein involved in modulation of the activity of the glucose-phosphotransferase system in Escherichia coli K-12.J Bacteriol. 2006 Aug;188(15):5439-49. doi: 10.1128/JB.00219-06. J Bacteriol. 2006. PMID: 16855233 Free PMC article.
-
The nature and dynamics of bacterial genomes.Science. 2006 Mar 24;311(5768):1730-3. doi: 10.1126/science.1119966. Science. 2006. PMID: 16556833 Review.
Cited by
-
Metabolic Profile of the Genome-Reduced Bacillus subtilis Strain IIG-Bs-27-39: An Attractive Chassis for Recombinant Protein Production.ACS Synth Biol. 2024 Jul 19;13(7):2199-2214. doi: 10.1021/acssynbio.4c00254. Epub 2024 Jul 9. ACS Synth Biol. 2024. PMID: 38981062 Free PMC article.
-
Group A streptococci induce stronger M protein-fibronectin interaction when specific human antibodies are bound.Front Microbiol. 2023 Jan 26;14:1069789. doi: 10.3389/fmicb.2023.1069789. eCollection 2023. Front Microbiol. 2023. PMID: 36778879 Free PMC article.
-
Evolutionary Plasticity of AmrZ Regulation in Pseudomonas.mSphere. 2018 Apr 18;3(2):e00132-18. doi: 10.1128/mSphere.00132-18. Print 2018 Apr 25. mSphere. 2018. PMID: 29669886 Free PMC article.
-
CRP improves the survival and competitive fitness of Salmonella Typhimurium under starvation by controlling the cellular maintenance rate.J Bacteriol. 2024 Aug 22;206(8):e0001024. doi: 10.1128/jb.00010-24. Epub 2024 Jul 24. J Bacteriol. 2024. PMID: 39046248 Free PMC article.
-
Whole genome-scale assessment of gene fitness of Novosphingobium aromaticavorans during spaceflight.BMC Genomics. 2023 Dec 16;24(1):782. doi: 10.1186/s12864-023-09799-z. BMC Genomics. 2023. PMID: 38102595 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

