The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing
- PMID: 27712628
- PMCID: PMC7112286
- DOI: 10.1016/bs.aivir.2016.08.008
The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing
Abstract
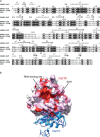
Coronaviruses are animal and human pathogens that can cause lethal zoonotic infections like SARS and MERS. They have polycistronic plus-stranded RNA genomes and belong to the order Nidovirales, a diverse group of viruses for which common ancestry was inferred from the common principles underlying their genome organization and expression, and from the conservation of an array of core replicase domains, including key RNA-synthesizing enzymes. Coronavirus genomes (~26-32 kilobases) are the largest RNA genomes known to date and their expansion was likely enabled by acquiring enzyme functions that counter the commonly high error frequency of viral RNA polymerases. The primary functions that direct coronavirus RNA synthesis and processing reside in nonstructural protein (nsp) 7 to nsp16, which are cleavage products of two large replicase polyproteins translated from the coronavirus genome. Significant progress has now been made regarding their structural and functional characterization, stimulated by technical advances like improved methods for bioinformatics and structural biology, in vitro enzyme characterization, and site-directed mutagenesis of coronavirus genomes. Coronavirus replicase functions include more or less universal activities of plus-stranded RNA viruses, like an RNA polymerase (nsp12) and helicase (nsp13), but also a number of rare or even unique domains involved in mRNA capping (nsp14, nsp16) and fidelity control (nsp14). Several smaller subunits (nsp7-nsp10) act as crucial cofactors of these enzymes and contribute to the emerging "nsp interactome." Understanding the structure, function, and interactions of the RNA-synthesizing machinery of coronaviruses will be key to rationalizing their evolutionary success and the development of improved control strategies.
Keywords: Capping; Coronavirus; Nidovirus; RNA processing; Replication.
© 2016 Elsevier Inc. All rights reserved.
Figures

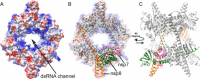




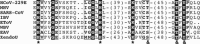
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

