Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig
- PMID: 27713410
- PMCID: PMC5059759
- DOI: 10.1038/ncomms13052
Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig
Abstract
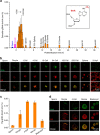
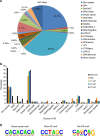
DNA N6-methyldeoxyadenosine (6mA) is a well-known prokaryotic DNA modification that has been shown to exist and play epigenetic roles in eukaryotic DNA. Here we report that 6mA accumulates up to ∼0.1-0.2% of total deoxyadenosine during early embryogenesis of vertebrates, but diminishes to the background level with the progression of the embryo development. During this process a large fraction of 6mAs locate in repetitive regions of the genome.
Figures
Similar articles
-
Epigenetic marking of the zebrafish developmental program.Curr Top Dev Biol. 2013;104:85-112. doi: 10.1016/B978-0-12-416027-9.00003-6. Curr Top Dev Biol. 2013. PMID: 23587239 Review.
-
Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm.Genome Res. 2011 Apr;21(4):578-89. doi: 10.1101/gr.113167.110. Epub 2011 Mar 7. Genome Res. 2011. PMID: 21383318 Free PMC article.
-
Embryonic DNA methylation: insights from the genomics era.Brief Funct Genomics. 2014 Mar;13(2):121-30. doi: 10.1093/bfgp/elt039. Epub 2013 Sep 23. Brief Funct Genomics. 2014. PMID: 24064195 Review.
-
Single-Nucleotide-Resolution Sequencing of N6-Methyldeoxyadenosine.Methods Mol Biol. 2021;2198:369-377. doi: 10.1007/978-1-0716-0876-0_28. Methods Mol Biol. 2021. PMID: 32822045
-
The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos.Methods. 2013 Aug 15;62(3):207-15. doi: 10.1016/j.ymeth.2013.04.011. Epub 2013 Apr 23. Methods. 2013. PMID: 23624103
Cited by
-
The Epigenetics of Gametes and Early Embryos and Potential Long-Range Consequences in Livestock Species-Filling in the Picture With Epigenomic Analyses.Front Genet. 2021 Mar 3;12:557934. doi: 10.3389/fgene.2021.557934. eCollection 2021. Front Genet. 2021. PMID: 33747031 Free PMC article. Review.
-
N6-Methyladenine DNA modification in Xanthomonas oryzae pv. oryzicola genome.Sci Rep. 2018 Nov 2;8(1):16272. doi: 10.1038/s41598-018-34559-5. Sci Rep. 2018. PMID: 30389999 Free PMC article.
-
Environmental Toxicity Assessment of Sodium Fluoride and Platinum-Derived Drugs Co-Exposure on Aquatic Organisms.Toxics. 2022 May 23;10(5):272. doi: 10.3390/toxics10050272. Toxics. 2022. PMID: 35622686 Free PMC article.
-
DNA N6-methyladenine in metazoans: functional epigenetic mark or bystander?Nat Struct Mol Biol. 2017 Jun 6;24(6):503-506. doi: 10.1038/nsmb.3412. Nat Struct Mol Biol. 2017. PMID: 28586322
-
Zebrafish transposable elements show extensive diversification in age, genomic distribution, and developmental expression.Genome Res. 2022 Jul;32(7):1408-1423. doi: 10.1101/gr.275655.121. Epub 2022 Jan 5. Genome Res. 2022. PMID: 34987056 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

