Bimodal antagonism of PKA signalling by ARHGAP36
- PMID: 27713425
- PMCID: PMC5059767
- DOI: 10.1038/ncomms12963
Bimodal antagonism of PKA signalling by ARHGAP36
Abstract
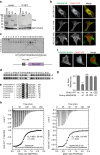
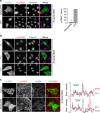
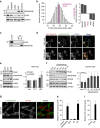
Protein kinase A is a key mediator of cAMP signalling downstream of G-protein-coupled receptors, a signalling pathway conserved in all eukaryotes. cAMP binding to the regulatory subunits (PKAR) relieves their inhibition of the catalytic subunits (PKAC). Here we report that ARHGAP36 combines two distinct inhibitory mechanisms to antagonise PKA signalling. First, it blocks PKAC activity via a pseudosubstrate motif, akin to the mechanism employed by the protein kinase inhibitor proteins. Second, it targets PKAC for rapid ubiquitin-mediated lysosomal degradation, a pathway usually reserved for transmembrane receptors. ARHGAP36 thus dampens the sensitivity of cells to cAMP. We show that PKA inhibition by ARHGAP36 promotes derepression of the Hedgehog signalling pathway, thereby providing a simple rationale for the upregulation of ARHGAP36 in medulloblastoma. Our work reveals a new layer of PKA regulation that may play an important role in development and disease.
Figures





References
-
- Cao Y. et al.. Activating hotspot L205R mutation in PRKACA and adrenal Cushing's syndrome. Science 344, 913–917 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

