The phasor-FLIM fingerprints reveal shifts from OXPHOS to enhanced glycolysis in Huntington Disease
- PMID: 27713486
- PMCID: PMC5054433
- DOI: 10.1038/srep34755
The phasor-FLIM fingerprints reveal shifts from OXPHOS to enhanced glycolysis in Huntington Disease
Abstract
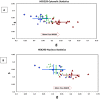
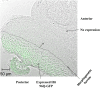
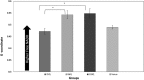
Huntington disease (HD) is an autosomal neurodegenerative disorder caused by the expansion of Polyglutamine (polyQ) in exon 1 of the Huntingtin protein. Glutamine repeats below 36 are considered normal while repeats above 40 lead to HD. Impairment in energy metabolism is a common trend in Huntington pathogenesis; however, this effect is not fully understood. Here, we used the phasor approach and Fluorescence Lifetime Imaging Microscopy (FLIM) to measure changes between free and bound fractions of NADH as a indirect measure of metabolic alteration in living cells. Using Phasor-FLIM, pixel maps of metabolic alteration in HEK293 cell lines and in transgenic Drosophila expressing expanded and unexpanded polyQ HTT exon1 in the eye disc were developed. We found a significant shift towards increased free NADH, indicating an increased glycolytic state for cells and tissues expressing the expanded polyQ compared to unexpanded control. In the nucleus, a further lifetime shift occurs towards higher free NADH suggesting a possible synergism between metabolic dysfunction and transcriptional regulation. Our results indicate that metabolic dysfunction in HD shifts to increased glycolysis leading to oxidative stress and cell death. This powerful label free method can be used to screen native HD tissue samples and for potential drug screening.
Figures



References
-
- Podolsky S., Leopold N. & Sax D. Increased frequency of diabetes mellitus in patients with Huntington’s chorea. The Lancet 299, 1356–1359 (1972). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

