Translation Elongation Factor eEF1A2 is a Novel Anticancer Target for the Marine Natural Product Plitidepsin
- PMID: 27713531
- PMCID: PMC5054363
- DOI: 10.1038/srep35100
Translation Elongation Factor eEF1A2 is a Novel Anticancer Target for the Marine Natural Product Plitidepsin
Abstract
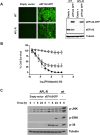
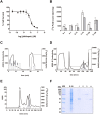
eEF1A2 is one of the isoforms of the alpha subunit of the eukaryotic Elongation Factor 1. It is overexpressed in human tumors and is endowed with oncogenic properties, favoring tumor cell proliferation while inhibiting apoptosis. We demonstrate that plitidepsin, an antitumor agent of marine origin that has successfully completed a phase-III clinical trial for multiple myeloma, exerts its antitumor activity by targeting eEF1A2. The drug interacts with eEF1A2 with a KD of 80 nM and a target residence time of circa 9 min. This protein was also identified as capable of binding [14C]-plitidepsin in a cell lysate from K-562 tumor cells. A molecular modelling approach was used to identify a favorable binding site for plitidepsin at the interface between domains 1 and 2 of eEF1A2 in the GTP conformation. Three tumor cell lines selected for at least 100-fold more resistance to plitidepsin than their respective parental cells showed reduced levels of eEF1A2 protein. Ectopic expression of eEF1A2 in resistant cells restored the sensitivity to plitidepsin. FLIM-phasor FRET experiments demonstrated that plitidepsin localizes in tumor cells sufficiently close to eEF1A2 as to suggest the formation of drug-protein complexes in living cells. Altogether, our results strongly suggest that eEF1A2 is the primary target of plitidepsin.
Conflict of interest statement
Alejandro Losada, María José Muñoz-Alonso, Juan Fernando Martínez-Leal, Juan Manuel Domínguez and Carlos M. Galmarini are employees and shareholders of Pharma Mar S.A. M.Pilar Lillo and Federico Gago have received a research grant from Pharma Mar S.A.
Figures



References
-
- Sun Y. et al.. Up-regulation of eEF1A2 promotes proliferation and inhibits apoptosis in prostate cancer. Biochem Biophys Res Commun 450, 1–6 (2014). - PubMed
-
- Hotokezaka Y. et al.. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J Biol Chem 277, 18545–18551 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

