In vitro characterization of biofilms formed by Kingella kingae
- PMID: 27714987
- PMCID: PMC5384882
- DOI: 10.1111/omi.12176
In vitro characterization of biofilms formed by Kingella kingae
Abstract
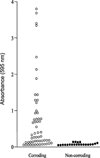
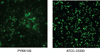
The Gram-negative bacterium Kingella kingae is part of the normal oropharyngeal mucosal flora of children <4 years old. K. kingae can enter the submucosa and cause infections of the skeletal system in children, including septic arthritis and osteomyelitis. The organism is also associated with infective endocarditis in children and adults. Although biofilm formation has been coupled with pharyngeal colonization, osteoarticular infections, and infective endocarditis, no studies have investigated biofilm formation in K. kingae. In this study we measured biofilm formation by 79 K. kingae clinical isolates using a 96-well microtiter plate crystal violet binding assay. We found that 37 of 79 strains (47%) formed biofilms. All strains that formed biofilms produced corroding colonies on agar. Biofilm formation was inhibited by proteinase K and DNase I. DNase I also caused the detachment of pre-formed K. kingae biofilm colonies. A mutant strain carrying a deletion of the pilus gene cluster pilA1pilA2fimB did not produce corroding colonies on agar, autoaggregate in broth, or form biofilms. Biofilm forming strains have higher levels of pilA1 expression. The extracellular components of biofilms contained 490 μg cm-2 of protein, 0.68 μg cm-2 of DNA, and 0.4 μg cm-2 of total carbohydrates. We concluded that biofilm formation is common among K. kingae clinical isolates, and that biofilm formation is dependent on the production of proteinaceous pili and extracellular DNA. Biofilm development may have relevance to the colonization, transmission, and pathogenesis of this bacterium. Extracellular DNA production by K. kingae may facilitate horizontal gene transfer within the oral microbial community.
Keywords: Kingella kingae; biofilms; oral microbiology.
© 2016 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures




References
-
- Amit U, Porat N, Basmaci R, et al. Genotyping of invasive Kingella kingae isolates reveals predominant clones and association with specific clinical syndromes. Clin Infect Dis. 2012;55:1074–1079. - PubMed
-
- Azakami H, Akimichi H, Usui M, Yumoto H, Ebisu S, Kato A. Isolation and characterization of a plasmid DNA from periodontopathogenic bacterium, Eikenella corrodens 1073, which affects pilus formation and colony morphology. Gene. 2005;351:143–148. - PubMed
-
- Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

