Assembly and trafficking of box C/D and H/ACA snoRNPs
- PMID: 27715451
- PMCID: PMC5519232
- DOI: 10.1080/15476286.2016.1243646
Assembly and trafficking of box C/D and H/ACA snoRNPs
Abstract
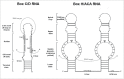
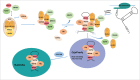
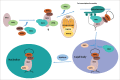
Box C/D and box H/ACA snoRNAs are abundant non-coding RNAs that localize in the nucleolus and mostly function as guides for nucleotide modifications. While a large pool of snoRNAs modifies rRNAs, an increasing number of snoRNAs could also potentially target mRNAs. ScaRNAs belong to a family of specific RNAs that localize in Cajal bodies and that are structurally similar to snoRNAs. Most scaRNAs are involved in snRNA modification, while telomerase RNA, which contains H/ACA motifs, functions in telomeric DNA synthesis. In this review, we describe how box C/D and H/ACA snoRNAs are processed and assembled with core proteins to form functional RNP particles. Their biogenesis involve several transport factors that first direct pre-snoRNPs to Cajal bodies, where some processing steps are believed to take place, and then to nucleoli. Assembly of core proteins involves the HSP90/R2TP chaperone-cochaperone system for both box C/D and H/ACA RNAs, but also several factors specific for each family. These assembly factors chaperone unassembled core proteins, regulate the formation and disassembly of pre-snoRNP intermediates, and control the activity of immature particles. The AAA+ ATPase RUVBL1 and RUVBL2 belong to the R2TP co-chaperones and play essential roles in snoRNP biogenesis, as well as in the formation of other macro-molecular complexes. Despite intensive research, their mechanisms of action are still incompletely understood.
Keywords: Hsp90/R2TP; RNP assembly; RNP trafficking; box C/D RNP; box H/ACA RNP; scaRNA; snoRNA.
Figures
References
-
- Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie 2002; 84:775-90; PMID:12457565; https://doi.org/10.1016/S0300-9084(02)01402-5 - DOI - PubMed
-
- Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr 2002; 10:17-39; PMID:11868985; https://doi.org/10.0000/096020197390059 - DOI - PMC - PubMed
-
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci 2002; 27:344-51; PMID:12114023; https://doi.org/10.1016/S0968-0004(02)02109-6 - DOI - PubMed
-
- Tollervey D, Hurt EC. The role of small nucleolar ribonucleoproteins in ribosome synthesis. Mol Biol Rep 1990; 14:103-6; PMID:2141891; https://doi.org/10.1007/BF00360433 - DOI - PubMed
-
- Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 1998; 2:629-38; PMID:9844635; https://doi.org/10.1016/S1097-2765(00)80161-6 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
