Putative RNA-directed adaptive mutations in cancer evolution
- PMID: 27715501
- PMCID: PMC5066521
- DOI: 10.1080/21541264.2016.1221491
Putative RNA-directed adaptive mutations in cancer evolution
Abstract
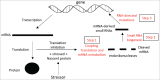
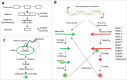
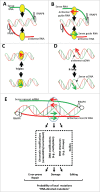
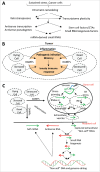
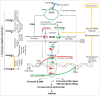
Understanding the molecular mechanisms behind the capacity of cancer cells to adapt to the tumor microenvironment and to anticancer therapies is a major challenge. In this context, cancer is believed to be an evolutionary process where random mutations and the selection process shape the mutational pattern and phenotype of cancer cells. This article challenges the notion of randomness of some cancer-associated mutations by describing molecular mechanisms involving stress-mediated biogenesis of mRNA-derived small RNAs able to target and increase the local mutation rate of the genomic loci they originate from. It is proposed that the probability of some mutations at specific loci could be increased in a stress-specific and RNA-depending manner. This would increase the probability of generating mutations that could alleviate stress situations, such as those triggered by anticancer drugs. Such a mechanism is made possible because tumor- and anticancer drug-associated stress situations trigger both cellular reprogramming and inflammation, which leads cancer cells to express molecular tools allowing them to "attack" and mutate their own genome in an RNA-directed manner.
Keywords: adaptive mutations; cancer; evolution; mutations; small RNAs.
Figures

Similar articles
-
Genome evolution is driven by gene expression-generated biophysical constraints through RNA-directed genetic variation: A hypothesis.Bioessays. 2017 Oct;39(10). doi: 10.1002/bies.201700069. Epub 2017 Sep 8. Bioessays. 2017. PMID: 28885715 Review.
-
The Role of Mutation Bias in Adaptive Evolution.Trends Ecol Evol. 2019 May;34(5):422-434. doi: 10.1016/j.tree.2019.01.015. Trends Ecol Evol. 2019. PMID: 31003616 Review.
-
Mutations, evolution and the central role of a self-defined fitness function in the initiation and progression of cancer.Biochim Biophys Acta Rev Cancer. 2017 Apr;1867(2):162-166. doi: 10.1016/j.bbcan.2017.03.005. Epub 2017 Mar 21. Biochim Biophys Acta Rev Cancer. 2017. PMID: 28341421 Free PMC article. Review.
-
Mutational randomness as conditional independence and the experimental vindication of mutational Lamarckism.Biol Rev Camb Philos Soc. 2017 May;92(2):673-683. doi: 10.1111/brv.12249. Epub 2016 Jan 29. Biol Rev Camb Philos Soc. 2017. PMID: 26824703
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
-
Overcoming randomness does not rule out the importance of inherent randomness for functionality.J Biosci. 2019 Dec;44(6):132. J Biosci. 2019. PMID: 31894113 Review.
-
Physicochemical Foundations of Life that Direct Evolution: Chance and Natural Selection are not Evolutionary Driving Forces.Life (Basel). 2020 Jan 21;10(2):7. doi: 10.3390/life10020007. Life (Basel). 2020. PMID: 31973071 Free PMC article.
References
-
- Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science 2015; 349:1483-1489. - PubMed
-
- Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: evolution within a lifetime. Annu Rev Genet 2014; 48:215-236. - PubMed
-
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science 2011; 331:1553-1558. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
