Novel microRNA discovery using small RNA sequencing in post-mortem human brain
- PMID: 27716130
- PMCID: PMC5050850
- DOI: 10.1186/s12864-016-3114-3
Novel microRNA discovery using small RNA sequencing in post-mortem human brain
Abstract
Background: MicroRNAs (miRNAs) are short, non-coding RNAs that regulate gene expression mainly through translational repression of target mRNA molecules. More than 2700 human miRNAs have been identified and some are known to be associated with disease phenotypes and to display tissue-specific patterns of expression.
Methods: We used high-throughput small RNA sequencing to discover novel miRNAs in 93 human post-mortem prefrontal cortex samples from individuals with Huntington's disease (n = 28) or Parkinson's disease (n = 29) and controls without neurological impairment (n = 36). A custom miRNA identification analysis pipeline was built, which utilizes miRDeep* miRNA identification and result filtering based on false positive rate estimates.
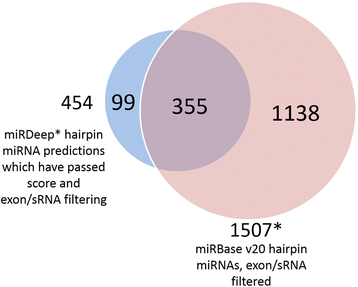
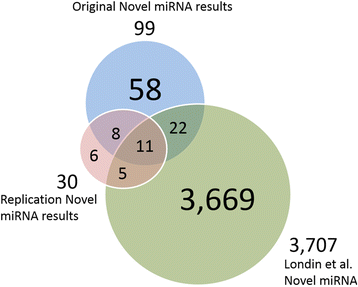
Results: Ninety-nine novel miRNA candidates with a false positive rate of less than 5 % were identified. Thirty-four of the candidate miRNAs show sequence similarity with known mature miRNA sequences and may be novel members of known miRNA families, while the remaining 65 may constitute previously undiscovered families of miRNAs. Nineteen of the 99 candidate miRNAs were replicated using independent, publicly-available human brain RNA-sequencing samples, and seven were experimentally validated using qPCR.
Conclusions: We have used small RNA sequencing to identify 99 putative novel miRNAs that are present in human brain samples.
Keywords: MicroRNA; Neurodegenerative disease; Novel miRNA discovery; Prefrontal cortex; miRNA sequencing.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

