Picodroplet partitioned whole genome amplification of low biomass samples preserves genomic diversity for metagenomic analysis
- PMID: 27716450
- PMCID: PMC5054601
- DOI: 10.1186/s40168-016-0197-7
Picodroplet partitioned whole genome amplification of low biomass samples preserves genomic diversity for metagenomic analysis
Abstract
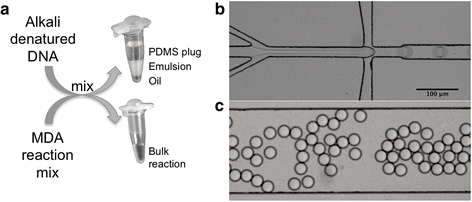
Background: Whole genome amplification (WGA) is a challenging, key step in metagenomic studies of samples containing minute amounts of DNA, such as samples from low biomass environments. It is well known that multiple displacement amplification (MDA), the most commonly used WGA method for microbial samples, skews the genomic representation in the sample. We have combined MDA with droplet microfluidics to perform the reaction in a homogeneous emulsion. Each droplet in this emulsion can be considered an individual reaction chamber, allowing partitioning of the MDA reaction into millions of parallel reactions with only one or very few template molecules per droplet.
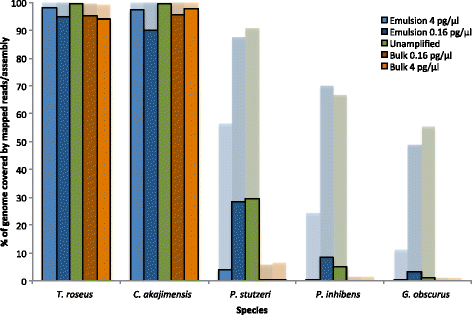
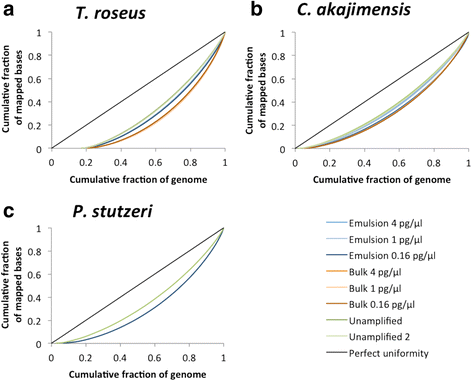
Results: As a proof-of-concept, we amplified genomic DNA from a synthetic metagenome by MDA either in one bulk reaction or in emulsion and found that after sequencing, the species distribution was better preserved and the coverage depth was more evenly distributed across the genomes when the MDA reaction had been performed in emulsion.
Conclusions: Partitioning MDA reactions into millions of reactions by droplet microfluidics is a straightforward way to improve the uniformity of MDA reactions for amplifying complex samples with limited amounts of DNA.
Keywords: Amplification bias; Droplet microfluidics; Metagenomics; Multiple displacement amplification; Whole genome amplification.
Figures

References
-
- Gonzalez JM, Portillo MC, Saiz-Jimenez C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ Microbiol. 2005;7:1024–8. doi: 10.1111/j.1462-2920.2005.00779.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

