Round Spermatid Injection Rescues Female Lethality of a Paternally Inherited Xist Deletion in Mouse
- PMID: 27716834
- PMCID: PMC5065126
- DOI: 10.1371/journal.pgen.1006358
Round Spermatid Injection Rescues Female Lethality of a Paternally Inherited Xist Deletion in Mouse
Abstract
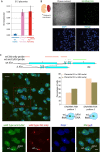
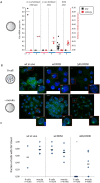
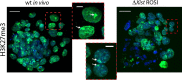
In mouse female preimplantation embryos, the paternal X chromosome (Xp) is silenced by imprinted X chromosome inactivation (iXCI). This requires production of the noncoding Xist RNA in cis, from the Xp. The Xist locus on the maternally inherited X chromosome (Xm) is refractory to activation due to the presence of an imprint. Paternal inheritance of an Xist deletion (XpΔXist) is embryonic lethal to female embryos, due to iXCI abolishment. Here, we circumvented the histone-to-protamine and protamine-to-histone transitions of the paternal genome, by fertilization of oocytes via injection of round spermatids (ROSI). This did not affect initiation of XCI in wild type female embryos. Surprisingly, ROSI using ΔXist round spermatids allowed survival of female embryos. This was accompanied by activation of the intact maternal Xist gene, initiated with delayed kinetics, around the morula stage, resulting in Xm silencing. Maternal Xist gene activation was not observed in ROSI-derived males. In addition, no Xist expression was detected in male and female morulas that developed from oocytes fertilized with mature ΔXist sperm. Finally, the expression of the X-encoded XCI-activator RNF12 was enhanced in both male (wild type) and female (wild type as well as XpΔXist) ROSI derived embryos, compared to in vivo fertilized embryos. Thus, high RNF12 levels may contribute to the specific activation of maternal Xist in XpΔXist female ROSI embryos, but upregulation of additional Xp derived factors and/or the specific epigenetic constitution of the round spermatid-derived Xp are expected to be more critical. These results illustrate the profound impact of a dysregulated paternal epigenome on embryo development, and we propose that mouse ROSI can be used as a model to study the effects of intergenerational inheritance of epigenetic marks.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

