Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid
- PMID: 27717386
- PMCID: PMC5054628
- DOI: 10.1186/s12934-016-0566-8
Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid
Abstract
Background: 5-Aminovaleric acid (5AVA) is an important five-carbon platform chemical that can be used for the synthesis of polymers and other chemicals of industrial interest. Enzymatic conversion of L-lysine to 5AVA has been achieved by employing lysine 2-monooxygenase encoded by the davB gene and 5-aminovaleramidase encoded by the davA gene. Additionally, a recombinant Escherichia coli strain expressing the davB and davA genes has been developed for bioconversion of L-lysine to 5AVA. To use glucose and xylose derived from lignocellulosic biomass as substrates, rather than L-lysine as a substrate, we previously examined direct fermentative production of 5AVA from glucose by metabolically engineered E. coli strains. However, the yield and productivity of 5AVA achieved by recombinant E. coli strains remain very low. Thus, Corynebacterium glutamicum, a highly efficient L-lysine producing microorganism, should be useful in the development of direct fermentative production of 5AVA using L-lysine as a precursor for 5AVA. Here, we report the development of metabolically engineered C. glutamicum strains for enhanced fermentative production of 5AVA from glucose.
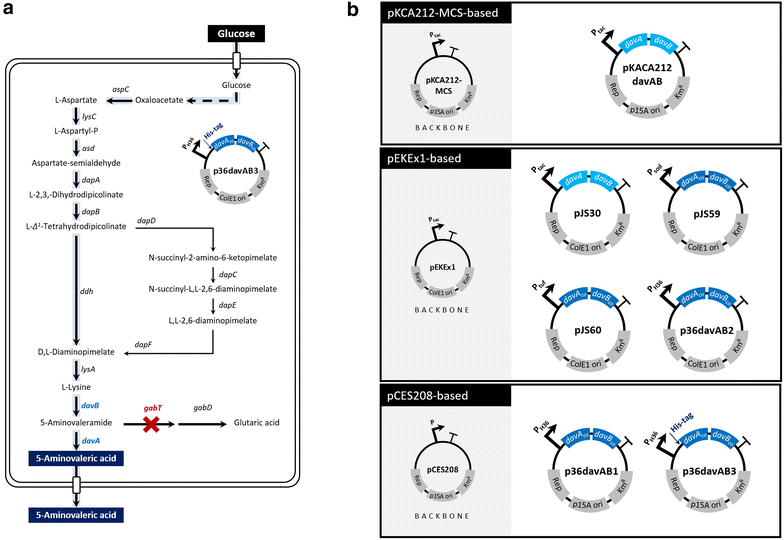
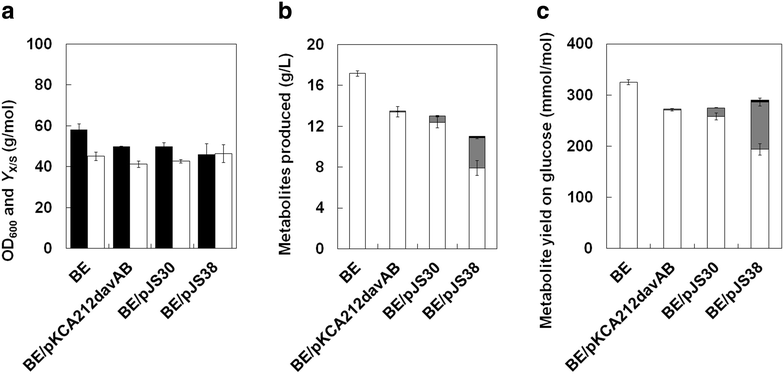
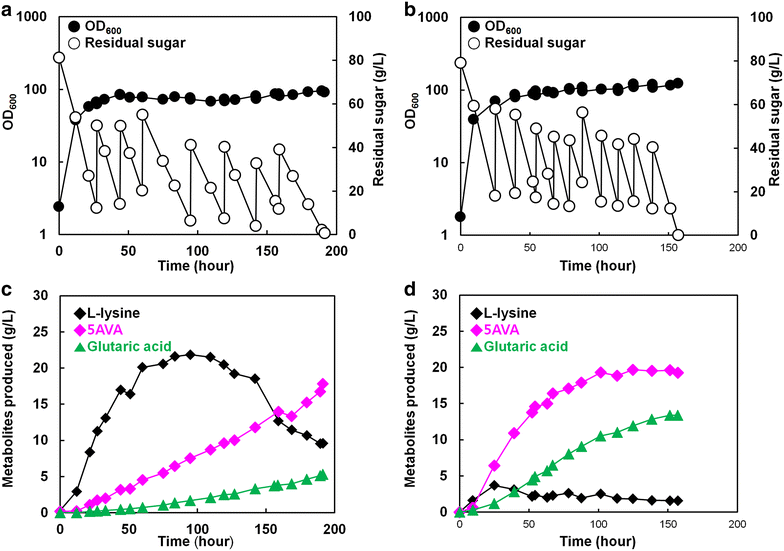
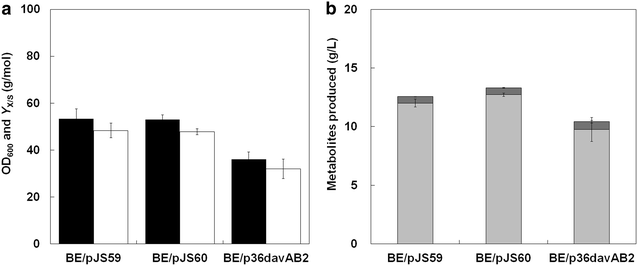
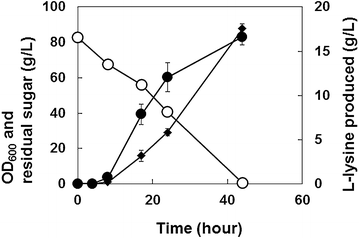
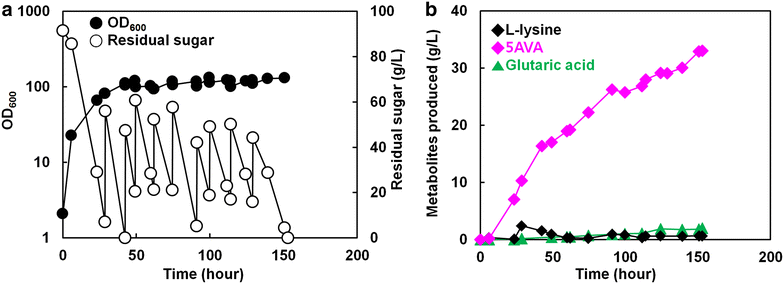
Results: Various expression vectors containing different promoters and origins of replication were examined for optimal expression of Pseudomonas putida davB and davA genes encoding lysine 2-monooxygenase and delta-aminovaleramidase, respectively. Among them, expression of the C. glutamicum codon-optimized davA gene fused with His6-Tag at its N-Terminal and the davB gene as an operon under a strong synthetic H36 promoter (plasmid p36davAB3) in C. glutamicum enabled the most efficient production of 5AVA. Flask culture and fed-batch culture of this strain produced 6.9 and 19.7 g/L (together with 11.9 g/L glutaric acid as major byproduct) of 5AVA, respectively. Homology modeling suggested that endogenous gamma-aminobutyrate aminotransferase encoded by the gabT gene might be responsible for the conversion of 5AVA to glutaric acid in recombinant C. glutamicum. Fed-batch culture of a C. glutamicum gabT mutant-harboring p36davAB3 produced 33.1 g/L 5AVA with much reduced (2.0 g/L) production of glutaric acid.
Conclusions: Corynebacterium glutamicum was successfully engineered to produce 5AVA from glucose by optimizing the expression of two key enzymes, lysine 2-monooxygenase and delta-aminovaleramidase. In addition, production of glutaric acid, a major byproduct, was significantly reduced by employing C. glutamicum gabT mutant as a host strain. The metabolically engineered C. glutamicum strains developed in this study should be useful for enhanced fermentative production of the novel C5 platform chemical 5AVA from renewable resources.
Keywords: 5-Aminovaleric acid; Corynebacterium glutamicum; Glutaric acid; L-Lysine; Metabolic engineering.
Figures

References
-
- Oh YH, Eom IY, Joo JC, Yu JH, Song BK, Lee SH, et al. Recent advances in development of biomass pretreatment technologies used in biorefinery for the production of bio-based fuels, chemicals and polymers. Korean J Chem Eng. 2015;32:1945–1959. doi: 10.1007/s11814-015-0191-y. - DOI
-
- Atabani AE, Silitonga AS, Badruddin IA, Mahlia TMI, Masjuki HH, Mekhilef S. A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew Sust Energ Rev. 2012;16:2070–2093. doi: 10.1016/j.rser.2012.01.003. - DOI
-
- Jang YS, Lee SY. Recent advances in biobutanol production. Industrial Biotechnol. 2015;11:316–321. doi: 10.1089/ind.2015.0023. - DOI
-
- Becker J, Klopprogge C, Zelder O, Heinzle E, Wittmann C. Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl Environ Microbiol. 2005;71:8587–8596. doi: 10.1128/AEM.71.12.8587-8596.2005. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

