Adaptation to Stressors by Systemic Protein Amyloidogenesis
- PMID: 27720612
- PMCID: PMC5098424
- DOI: 10.1016/j.devcel.2016.09.002
Adaptation to Stressors by Systemic Protein Amyloidogenesis
Abstract
The amyloid state of protein organization is typically associated with debilitating human neuropathies and is seldom observed in physiology. Here, we uncover a systemic program that leverages the amyloidogenic propensity of proteins to regulate cell adaptation to stressors. On stimulus, cells assemble the amyloid bodies (A-bodies), nuclear foci containing heterogeneous proteins with amyloid-like biophysical properties. A discrete peptidic sequence, termed the amyloid-converting motif (ACM), is capable of targeting proteins to the A-bodies by interacting with ribosomal intergenic noncoding RNA (rIGSRNA). The pathological β-amyloid peptide, involved in Alzheimer's disease, displays ACM-like activity and undergoes stimuli-mediated amyloidogenesis in vivo. Upon signal termination, elements of the heat-shock chaperone pathway disaggregate the A-bodies. Physiological amyloidogenesis enables cells to store large quantities of proteins and enter a dormant state in response to stressors. We suggest that cells have evolved a post-translational pathway that rapidly and reversibly converts native-fold proteins to an amyloid-like solid phase.
Keywords: Hsp70; amyloid-bodies (A-bodies); dormancy; extracellular stress; heat shock chaperones; long noncoding RNA (lncRNA); physiological amyloidogenesis; β-amyloid.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

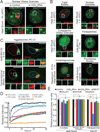


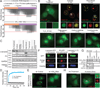

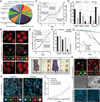
Comment in
-
RNA-Seeded Functional Amyloids Balance Growth and Survival.Dev Cell. 2016 Oct 24;39(2):131-132. doi: 10.1016/j.devcel.2016.10.005. Dev Cell. 2016. PMID: 27780035 Free PMC article.
References
-
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. - PubMed
-
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. - PubMed
-
- Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–14163. - PubMed
-
- Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, Shepard HM, Osborne CK. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24:85–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

