RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation
- PMID: 27720642
- PMCID: PMC5107313
- DOI: 10.1016/j.molcel.2016.08.035
RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation
Abstract
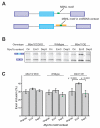
Many RNA binding proteins (RBPs) bind specific RNA sequence motifs, but only a small fraction (∼15%-40%) of RBP motif occurrences are occupied in vivo. To determine which contextual features discriminate between bound and unbound motifs, we performed an in vitro binding assay using 12,000 mouse RNA sequences with the RBPs MBNL1 and RBFOX2. Surprisingly, the strength of binding to motif occurrences in vitro was significantly correlated with in vivo binding, developmental regulation, and evolutionary age of alternative splicing. Multiple lines of evidence indicate that the primary context effect that affects binding in vitro and in vivo is RNA secondary structure. Large-scale combinatorial mutagenesis of unfavorable sequence contexts revealed a consistent pattern whereby mutations that increased motif accessibility improved protein binding and regulatory activity. Our results indicate widespread inhibition of motif binding by local RNA secondary structure and suggest that mutations that alter sequence context commonly affect RBP binding and regulation.
Keywords: RBNS; RNA processing; post-transcriptional.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


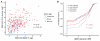



Comment in
-
RNA Structure Switches RBP Binding.Mol Cell. 2016 Oct 20;64(2):219-220. doi: 10.1016/j.molcel.2016.10.006. Mol Cell. 2016. PMID: 27768870
Similar articles
-
RNA Bind-n-Seq: Measuring the Binding Affinity Landscape of RNA-Binding Proteins.Methods Enzymol. 2015;558:465-493. doi: 10.1016/bs.mie.2015.02.007. Epub 2015 May 12. Methods Enzymol. 2015. PMID: 26068750 Free PMC article.
-
A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data.Methods. 2017 Apr 15;118-119:73-81. doi: 10.1016/j.ymeth.2017.03.003. Epub 2017 Mar 6. Methods. 2017. PMID: 28274760
-
Prediction of clustered RNA-binding protein motif sites in the mammalian genome.Nucleic Acids Res. 2013 Aug;41(14):6793-807. doi: 10.1093/nar/gkt421. Epub 2013 May 18. Nucleic Acids Res. 2013. PMID: 23685613 Free PMC article.
-
Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition.Methods. 2017 Apr 15;118-119:119-136. doi: 10.1016/j.ymeth.2017.03.015. Epub 2017 Mar 16. Methods. 2017. PMID: 28315749 Review.
-
RNA-protein interactions: an overview.Methods Mol Biol. 2014;1097:491-521. doi: 10.1007/978-1-62703-709-9_23. Methods Mol Biol. 2014. PMID: 24639174 Review.
Cited by
-
Artificial intelligence methods enhance the discovery of RNA interactions.Front Mol Biosci. 2022 Oct 7;9:1000205. doi: 10.3389/fmolb.2022.1000205. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275611 Free PMC article. Review.
-
Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches.Nat Commun. 2023 Feb 10;14(1):752. doi: 10.1038/s41467-023-36316-3. Nat Commun. 2023. PMID: 36765063 Free PMC article.
-
Structural disruption of exonic stem-loops immediately upstream of the intron regulates mammalian splicing.Nucleic Acids Res. 2020 Jun 19;48(11):6294-6309. doi: 10.1093/nar/gkaa358. Nucleic Acids Res. 2020. PMID: 32402057 Free PMC article.
-
RNA binding proteins (RBPs) on genetic stability and diseases.Glob Med Genet. 2024 Nov 30;12(1):100032. doi: 10.1016/j.gmg.2024.100032. eCollection 2025 Mar. Glob Med Genet. 2024. PMID: 39925443 Free PMC article. Review.
-
Denaturing CLIP, dCLIP, Pipeline Identifies Discrete RNA Footprints on Chromatin-Associated Proteins and Reveals that CBX7 Targets 3' UTRs to Regulate mRNA Expression.Cell Syst. 2017 Oct 25;5(4):368-385.e15. doi: 10.1016/j.cels.2017.09.014. Cell Syst. 2017. PMID: 29073373 Free PMC article.
References
-
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
