Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process
- PMID: 27720644
- PMCID: PMC5123691
- DOI: 10.1016/j.molcel.2016.09.005
Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process
Abstract
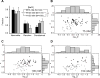
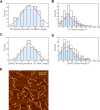
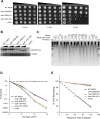
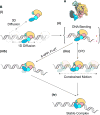
Nucleotide excision repair (NER) is an evolutionarily conserved mechanism that processes helix-destabilizing and/or -distorting DNA lesions, such as UV-induced photoproducts. Here, we investigate the dynamic protein-DNA interactions during the damage recognition step using single-molecule fluorescence microscopy. Quantum dot-labeled Rad4-Rad23 (yeast XPC-RAD23B ortholog) forms non-motile complexes or conducts a one-dimensional search via either random diffusion or constrained motion. Atomic force microcopy analysis of Rad4 with the β-hairpin domain 3 (BHD3) deleted reveals that this motif is non-essential for damage-specific binding and DNA bending. Furthermore, we find that deletion of seven residues in the tip of β-hairpin in BHD3 increases Rad4-Rad23 constrained motion at the expense of stable binding at sites of DNA lesions, without diminishing cellular UV resistance or photoproduct repair in vivo. These results suggest a distinct intermediate in the damage recognition process during NER, allowing dynamic DNA damage detection at a distance.
Keywords: DNA tightrope assay; Rad23; Rad4; XPC; dynamic DNA damage recognition; nucleotide excision repair; quantum dots; single particle tracking; xeroderma pigmentosum.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. - PubMed
-
- Brown KL, Roginskaya M, Zou Y, Altamirano A, Basu AK, Stone MP. Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5′-GTgG-3′ sequence: destabilization of two base pairs at the lesion site. Nucleic acids research. 2010;38:428–440. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

