Pairing beyond the Seed Supports MicroRNA Targeting Specificity
- PMID: 27720646
- PMCID: PMC5074850
- DOI: 10.1016/j.molcel.2016.09.004
Pairing beyond the Seed Supports MicroRNA Targeting Specificity
Abstract
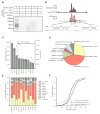
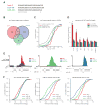
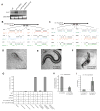
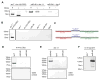
To identify endogenous miRNA-target sites, we isolated AGO-bound RNAs from Caenorhabditis elegans by individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP), which fortuitously also produced miRNA-target chimeric reads. Through the analysis of thousands of reproducible chimeras, pairing to the miRNA seed emerged as the predominant motif associated with functional interactions. Unexpectedly, we discovered that additional pairing to 3' sequences is prevalent in the majority of target sites and leads to specific targeting by members of miRNA families. By editing an endogenous target site, we demonstrate that 3' pairing determines targeting by specific miRNA family members and that seed pairing is not always sufficient for functional target interactions. Finally, we present a simplified method, chimera PCR (ChimP), for the detection of specific miRNA-target interactions. Overall, our analysis revealed that sequences in the 5' as well as the 3' regions of a miRNA provide the information necessary for stable and specific miRNA-target interactions in vivo.
Keywords: ALG-1; C. elegans; chimeras; iCLIP; miRNA family; microRNA.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

