Genomics-assisted characterization of a breeding collection of Apios americana, an edible tuberous legume
- PMID: 27721469
- PMCID: PMC5056515
- DOI: 10.1038/srep34908
Genomics-assisted characterization of a breeding collection of Apios americana, an edible tuberous legume
Abstract
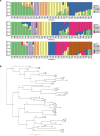
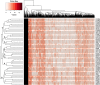
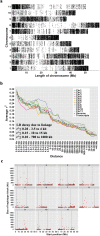
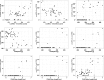
For species with potential as new crops, rapid improvement may be facilitated by new genomic methods. Apios (Apios americana Medik.), once a staple food source of Native American Indians, produces protein-rich tubers, tolerates a wide range of soils, and symbiotically fixes nitrogen. We report the first high-quality de novo transcriptome assembly, an expression atlas, and a set of 58,154 SNP and 39,609 gene expression markers (GEMs) for characterization of a breeding collection. Both SNPs and GEMs identify six genotypic clusters in the collection. Transcripts mapped to the Phaseolus vulgaris genome-another phaseoloid legume with the same chromosome number-provide provisional genetic locations for 46,852 SNPs. Linkage disequilibrium decays within 10 kb (based on the provisional genetic locations), consistent with outcrossing reproduction. SNPs and GEMs identify more than 21 marker-trait associations for at least 11 traits. This study demonstrates a holistic approach for mining plant collections to accelerate crop improvement.
Figures


References
-
- Vega-Galvez A. et al.. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: a review. J Sci Food Agric 90, 2541–2547 (2010). - PubMed
-
- Peterson A., Jacobsen S.-E., Bonifacio A. & Murphy K. A Crossing Method for Quinoa. Sustainability 7, 3230–3243 (2015).
-
- Wu G., Morris C. F. & Murphy K. M. Evaluation of texture differences among varieties of cooked quinoa. J Food Sci 79, S2337–S2345 (2014). - PubMed
-
- Cox T. S., Van Tassel D. L., Cox C. M. & DeHaan L. R. Progress in breeding perennial grains. Crop and Pasture Science 61, 513–521 (2010).
-
- DeHaan L. R. Progress in developing Kernza wheatgrass as a perennial grain. Water, Food, Energy & Innovation for a Sustainable World, Annual Report of The Land Institute, https://landinstitute.org/sites/default/files/fy2014_annual_report.pdf (2014).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

