Components of Adenovirus Genome Packaging
- PMID: 27721809
- PMCID: PMC5033970
- DOI: 10.3389/fmicb.2016.01503
Components of Adenovirus Genome Packaging
Abstract
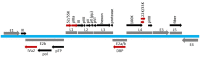
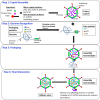
Adenoviruses (AdVs) are icosahedral viruses with double-stranded DNA (dsDNA) genomes. Genome packaging in AdV is thought to be similar to that seen in dsDNA containing icosahedral bacteriophages and herpesviruses. Specific recognition of the AdV genome is mediated by a packaging domain located close to the left end of the viral genome and is mediated by the viral packaging machinery. Our understanding of the role of various components of the viral packaging machinery in AdV genome packaging has greatly advanced in recent years. Characterization of empty capsids assembled in the absence of one or more components involved in packaging, identification of the unique vertex, and demonstration of the role of IVa2, the putative packaging ATPase, in genome packaging have provided compelling evidence that AdVs follow a sequential assembly pathway. This review provides a detailed discussion on the functions of the various viral and cellular factors involved in AdV genome packaging. We conclude by briefly discussing the roles of the empty capsids, assembly intermediates, scaffolding proteins, portal vertex and DNA encapsidating enzymes in AdV assembly and packaging.
Keywords: ATPase; IVa2; L4 22K; L4 33K; adenovirus; genome packaging; packaging domain; portal vertex.
Figures


References
-
- Ariga H., Klein H., Levine A. J., Horwitz M. S. (1980). A cleavage product of the adenovirus DNA binding protein is active in DNA replication in vitro. Virology 101, 307–310. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

