Genome-Wide Identification of BAHD Acyltransferases and In vivo Characterization of HQT-like Enzymes Involved in Caffeoylquinic Acid Synthesis in Globe Artichoke
- PMID: 27721818
- PMCID: PMC5033976
- DOI: 10.3389/fpls.2016.01424
Genome-Wide Identification of BAHD Acyltransferases and In vivo Characterization of HQT-like Enzymes Involved in Caffeoylquinic Acid Synthesis in Globe Artichoke
Abstract
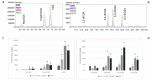
Globe artichoke (Cynara cardunculus L. var. scolymus) is a rich source of compounds promoting human health (phytonutrients), among them caffeoylquinic acids (CQAs), mainly represented by chlorogenic acid (CGA), and dicaffeoylquinic acids (diCQAs). The enzymes involved in their biosynthesis belong to the large family of BAHD acyltransferases. Following a survey of the globe artichoke genome, we identified 69 BAHD proteins carrying the catalytic site (HXXXD). Their phylogenetic analysis together with another 43 proteins, from 21 species, representative of the BAHD family, highlighted their grouping in seven major clades. Nine globe artichoke acyltransferases clustered in a sub-group of Clade V, with 3 belonging to hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferase (HQT) and 2 to hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT) like proteins. We focused our attention on the former, HQT1, HQT2, and HQT3, as they are known to play a key role in CGA biosynthesis. The expression of genes coding for the three HQTs and correlation of expression with the CQA content is reported for different globe artichoke tissues. For the first time in the globe artichoke, we developed and applied the virus-induced gene silencing approach with the goal of assessing in vivo the effect of HQT1 silencing, which resulted in a marked reduction of both CGA and diCQAs. On the other hand, when the role of the three HQTs was assessed in leaves of Nicotiana benthamiana through their transient overexpression, significant increases in mono- and diCQAs content were observed. Using transient GFP fusion proteins expressed in N. benthamiana leaves we also established the sub-cellular localization of these three enzymes.
Keywords: BAHD acyltransferases; Cynara cardunculus; VIGS; caffeoylquinic acids; functional characterization.
Figures







References
-
- Bai S., Tuan P., Tatsuki M., Yaegaki H., Ohmiya A., Yamamizo C., et al. (2016). Knockdown of carotenoid cleavage dioxygenase 4 (CCD4) via virus-induced gene silencing confers yellow coloration in peach fruit: evaluation of gene function related to fruit traits. Plant Mol. Biol. Rep. 34 257–264. 10.1007/s11105-015-0920-8 - DOI
-
- Barker M., Kane N., Matvienko M., Kozik A., Michelmore W., Knapp S., et al. (2008). Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 25 2445–2455. 10.1093/molbev/msn187 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

