Pathogenic infection of Rhesus macaques by an evolving SIV-HIV derived from CCR5-using envelope genes of acute HIV-1 infections
- PMID: 27723488
- PMCID: PMC5899904
- DOI: 10.1016/j.virol.2016.09.021
Pathogenic infection of Rhesus macaques by an evolving SIV-HIV derived from CCR5-using envelope genes of acute HIV-1 infections
Abstract
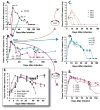
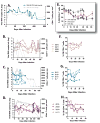
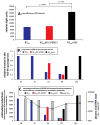
For studies on vaccines and therapies for HIV disease, SIV-HIV chimeric viruses harboring the HIV-1 env gene (SHIVenv) remain the best virus in non-human primate models. However, there are still very few SHIVenv viruses that can cause AIDS in non-CD8-depleted animals. In the present study, a recently created CCR5-using SHIVenv_B3 virus with env gene derived from acute/early HIV-1 infections (AHI) successfully established pathogenic infection in macaques. Through a series of investigations on the evolution, mutational profile, and phenotype of the virus and the resultant humoral immune response in infected rhesus macaques, we found that the E32K mutation in the Env C1 domain was associated with macaque pathogenesis, and that the electrostatic interactions in Env may favor E32K at the gp120 N terminus and "lock" the binding to heptad repeat 1 of gp41 in the trimer and produce a SHIVenv with increased fitness and pathogenesis during macaque infections.
Keywords: Envelope; Pathogenesis; Rhesus macaque; SIV-HIV chimeric viruses.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Infection of rhesus macaques with a pool of simian immunodeficiency virus with the envelope genes from acute HIV-1 infections.AIDS Res Ther. 2016 Nov 25;13(1):41. doi: 10.1186/s12981-016-0125-8. AIDS Res Ther. 2016. PMID: 27906032 Free PMC article.
-
Bovine alpha-2-HS-glycoprotein functions as a booster antigen for efficiently stimulating humoral immune responses to CCR5 and SIVmac239 envelope glycoprotein.Biochem Biophys Res Commun. 2014 Jan 3;443(1):301-7. doi: 10.1016/j.bbrc.2013.11.098. Epub 2013 Dec 2. Biochem Biophys Res Commun. 2014. PMID: 24309114
-
Chronology of genetic changes in the vpu, env, and Nef genes of chimeric simian-human immunodeficiency virus (strain HXB2) during acquisition of virulence for pig-tailed macaques.Virology. 1998 Sep 1;248(2):275-83. doi: 10.1006/viro.1998.9300. Virology. 1998. PMID: 9721236
-
Minimally Modified HIV-1 Infection of Macaques: Development, Utility, and Limitations of Current Models.Viruses. 2024 Oct 16;16(10):1618. doi: 10.3390/v16101618. Viruses. 2024. PMID: 39459950 Free PMC article. Review.
-
Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence.Curr Top Microbiol Immunol. 1994;188:185-219. doi: 10.1007/978-3-642-78536-8_11. Curr Top Microbiol Immunol. 1994. PMID: 7523031 Review.
Cited by
-
Identification of HIV-1 Envelope Mutations that Enhance Entry Using Macaque CD4 and CCR5.Viruses. 2020 Feb 21;12(2):241. doi: 10.3390/v12020241. Viruses. 2020. PMID: 32098152 Free PMC article.
-
Early phase dynamics of traceable mCherry fluorescent protein-carrying HIV-1 infection in human peripheral blood mononuclear cells-transplanted NOD/SCID/Jak3-/- mice.Antiviral Res. 2017 Aug;144:83-92. doi: 10.1016/j.antiviral.2017.03.027. Epub 2017 Apr 7. Antiviral Res. 2017. PMID: 28392419 Free PMC article.
-
Frequency of Human CD45+ Target Cells is a Key Determinant of Intravaginal HIV-1 Infection in Humanized Mice.Sci Rep. 2017 Nov 10;7(1):15263. doi: 10.1038/s41598-017-15630-z. Sci Rep. 2017. PMID: 29127409 Free PMC article.
References
-
- Barnett SW, Burke B, Sun Y, Kan E, Legg H, Lian Y, Bost K, Zhou F, Goodsell A, zur MJ, Polo J, Donnelly J, Ulmer J, Otten GR, Miller CJ, Vajdy M, Srivastava IK. Antibody-mediated protection against mucosal simian-human immunodeficiency virus challenge of macaques immunized with alphavirus replicon particles and boosted with trimeric envelope glycoprotein in MF59 adjuvant. J Virol. 2010;84:5975–5985. - PMC - PubMed
-
- Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim JH, Korber BT, Michael NL. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539. - PMC - PubMed
-
- Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. - PMC - PubMed
-
- Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho DD. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina) J Virol. 2000;74:6501–6510. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

