Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants
- PMID: 27723757
- PMCID: PMC5120758
- DOI: 10.1038/ng.3680
Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants
Abstract
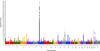
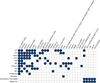
Vitiligo is an autoimmune disease in which depigmented skin results from the destruction of melanocytes, with epidemiological association with other autoimmune diseases. In previous linkage and genome-wide association studies (GWAS1 and GWAS2), we identified 27 vitiligo susceptibility loci in patients of European ancestry. We carried out a third GWAS (GWAS3) in European-ancestry subjects, with augmented GWAS1 and GWAS2 controls, genome-wide imputation, and meta-analysis of all three GWAS, followed by an independent replication. The combined analyses, with 4,680 cases and 39,586 controls, identified 23 new significantly associated loci and 7 suggestive loci. Most encode immune and apoptotic regulators, with some also associated with other autoimmune diseases, as well as several melanocyte regulators. Bioinformatic analyses indicate a predominance of causal regulatory variation, some of which corresponds to expression quantitative trait loci (eQTLs) at these loci. Together, the identified genes provide a framework for the genetic architecture and pathobiology of vitiligo, highlight relationships with other autoimmune diseases and melanoma, and offer potential targets for treatment.
Figures

References
-
- Picardo M, Taïeb A, editors. Vitiligo. Springer, Heidelberg & New York: 2010.
-
- Alkhateeb A, et al. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. - PubMed
-
- Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 2007;356:1216–1225. - PubMed
Methods-only References
-
- Taïeb A, Picardo M. The definition and assessment of vitiligo: a consensus report of the VitiligoEuropean Task Force. Pigment Cell Res. 2007;20:27–35. - PubMed
-
- Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

