A Tale of Tails: Dissecting the Enhancing Effect of Tailed Primers in Real-Time PCR
- PMID: 27723800
- PMCID: PMC5056738
- DOI: 10.1371/journal.pone.0164463
A Tale of Tails: Dissecting the Enhancing Effect of Tailed Primers in Real-Time PCR
Abstract
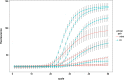
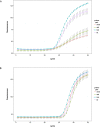
Non-specific tail sequences are often added to the 5'-terminus of primers to improve the robustness and overall performance of diagnostic assays. Despite the widespread use of tailed primers, the underlying working mechanism is not well understood. To address this problem, we conducted a detailed in vitro and in silico analysis of the enhancing effect of primer tailing on 2 well-established foot-and-mouth disease virus (FMDV) RT-qPCR assays using an FMDV reference panel. Tailing of the panFMDV-5UTR primers mainly affected the shape of the amplification curves. Modelling of the raw fluorescence data suggested a reduction of the amplification efficiency due to the accumulation of inhibitors. In depth analysis of PCR products indeed revealed the rapid accumulation of forward-primer derived artefacts. More importantly, tailing of the forward primer delayed artefacts formation and concomitantly restored the sigmoidal shape of the amplification curves. Our analysis also showed that primer tailing can alter utilisation patterns of degenerate primers and increase the number of primer variants that are able to participate in the reaction. The impact of tailed primers was less pronounced in the panFMDV-3D assay with only 5 out of 50 isolates showing a clear shift in Cq values. Sequence analysis of the target region of these 5 isolates revealed several mutations in the inter-primer region that extend an existing hairpin structure immediately downstream of the forward primer binding site. Stabilisation of the forward primer with either a tail sequence or cationic spermine units restored the sensitivity of the assay, which suggests that the enhancing effect in the panFMDV-3D assay is due to a more efficient extension of the forward primer. ur results show that primer tailing can alter amplification through various mechanisms that are determined by both the assay and target region. These findings expand our understanding of primer tailing and should enable a more targeted and efficient use of tailed primers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kaufman DL, Evans GA. Restriction endonuclease cleavage at the termini of PCR products. Biotechniques. 1990; 9(3): 304, 306. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

