A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome
- PMID: 27725671
- PMCID: PMC5062574
- DOI: 10.1038/ncomms12522
A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome
Abstract
The African Diaspora in the Western Hemisphere represents one of the largest forced migrations in history and had a profound impact on genetic diversity in modern populations. To date, the fine-scale population structure of descendants of the African Diaspora remains largely uncharacterized. Here we present genetic variation from deeply sequenced genomes of 642 individuals from North and South American, Caribbean and West African populations, substantially increasing the lexicon of human genomic variation and suggesting much variation remains to be discovered in African-admixed populations in the Americas. We summarize genetic variation in these populations, quantifying the postcolonial sex-biased European gene flow across multiple regions. Moreover, we refine estimates on the burden of deleterious variants carried across populations and how this varies with African ancestry. Our data are an important resource for empowering disease mapping studies in African-admixed individuals and will facilitate gene discovery for diseases disproportionately affecting individuals of African ancestry.
Conflict of interest statement
Nadia N. Hansel has a consulting relationship with GSK (Advisory Board). All other authors declare no competing financial interests.
Figures

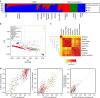
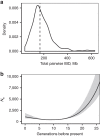

References
-
- Tishkoff S. A. & Kidd K. K. Implications of biogeography of human populations for 'race' and medicine. Nat. Genet. 36, S21–S27 (2004). - PubMed
-
- Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA). http://www.caapaproject.org (2012).
-
- Duffy D. L., Martin N. G., Battistutta D., Hopper J. L. & Mathews J. D. Genetics of asthma and hay fever in Australian twins. Am. Rev. Resp. Dis. 142, 1351–1358 (1990). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- K24 AI077930/AI/NIAID NIH HHS/United States
- G12 RR003048/RR/NCRR NIH HHS/United States
- P50 CA125183/CA/NCI NIH HHS/United States
- U01 CA161032/CA/NCI NIH HHS/United States
- RC2 HL101543/HL/NHLBI NIH HHS/United States
- R01 AI079139/AI/NIAID NIH HHS/United States
- HHSN268201300048C/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- R01 HL087699/HL/NHLBI NIH HHS/United States
- R21 HG004751/HG/NHGRI NIH HHS/United States
- U01 HL049596/HL/NHLBI NIH HHS/United States
- U19 AI095227/AI/NIAID NIH HHS/United States
- R37 HL068546/HL/NHLBI NIH HHS/United States
- P30 DK020595/DK/NIDDK NIH HHS/United States
- R01 HL069167/HL/NHLBI NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- U54 MD007588/MD/NIMHD NIH HHS/United States
- K23 HL004464/HL/NHLBI NIH HHS/United States
- R21 HG007233/HG/NHGRI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- R21 CA178706/CA/NCI NIH HHS/United States
- U19 AI095230/AI/NIAID NIH HHS/United States
- R01 HG007644/HG/NHGRI NIH HHS/United States
- R01 AI114555/AI/NIAID NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- RC2 HL101651/HL/NHLBI NIH HHS/United States
- HHSN268201300049C/HL/NHLBI NIH HHS/United States
- HHSN268201300047C/HL/NHLBI NIH HHS/United States
- R01 HL104608/HL/NHLBI NIH HHS/United States
- HHSN268201300050C/HL/NHLBI NIH HHS/United States
- U10 HL109164/HL/NHLBI NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- P60 MD006902/MD/NIMHD NIH HHS/United States
- R01 HL072414/HL/NHLBI NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- HHSN268201300046C/HL/NHLBI NIH HHS/United States
- S06 GM008016/GM/NIGMS NIH HHS/United States
- K01 HL092601/HL/NHLBI NIH HHS/United States
- R21 HL112656/HL/NHLBI NIH HHS/United States
- R01 HL088133/HL/NHLBI NIH HHS/United States
- U01 HL081332/HL/NHLBI NIH HHS/United States
- T32 GM007175/GM/NIGMS NIH HHS/United States
- R01 HL118267/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

