Microbiome and Exudates of the Root and Rhizosphere of Brachypodium distachyon, a Model for Wheat
- PMID: 27727301
- PMCID: PMC5058512
- DOI: 10.1371/journal.pone.0164533
Microbiome and Exudates of the Root and Rhizosphere of Brachypodium distachyon, a Model for Wheat
Abstract
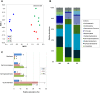
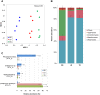
The rhizosphere microbiome is regulated by plant genotype, root exudates and environment. There is substantial interest in breeding and managing crops that host root microbial communities that increase productivity. The eudicot model species Arabidopsis has been used to investigate these processes, however a model for monocotyledons is also required. We characterized the rhizosphere microbiome and root exudates of Brachypodium distachyon, to develop it as a rhizosphere model for cereal species like wheat. The Brachypodium rhizosphere microbial community was dominated by Burkholderiales. However, these communities were also dependent on how tightly they were bound to roots, the root type they were associated with (nodal or seminal roots), and their location along the roots. Moreover, the functional gene categories detected in microorganisms isolated from around root tips differed from those isolated from bases of roots. The Brachypodium rhizosphere microbiota and root exudate profiles were similar to those reported for wheat rhizospheres, and different to Arabidopsis. The differences in root system development and cell wall chemistry between monocotyledons and eudicots may also influence the microorganism composition of these major plant types. Brachypodium is a promising model for investigating the microbiome of wheat.
Conflict of interest statement
The authors have declared that no competing interests exist. Please note that the Forschungszentrum Juelich is not a private entity. It is a public, not-for-profit research centre focusing on societal research needs (with majority public funds, and these from German Federal Government (90%) and State Government (10%)) (http://www.fz-juelich.de/portal/EN/AboutUs/FactsFigures/_node.html), and is a member of the Helmholtz Association of German research centres, Germany's largest public research association dedicated to research for state and society (https://www.helmholtz.de/en/about_us/).
Figures





References
-
- Vogel JP, Garvin DF, Leong OM, Hayden DM. Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult. 2006;84(2):199–211. 10.1007/s11240-005-9023-9. WOS:000237967000008. - DOI
-
- Budak H, Hernandez P, Schulman HA. Analysis and exploitation of cereal genomes with the aid of Brachypodium In: Tuberosa R, Graner A, Frison E, editors. Genomics of plant genetic resources: Volume 1 Managing, sequencing and mining genetic resources. Dordrecht: Springer Netherlands; 2014. p. 585–613. 10.1007/978-94-007-7572-5_24 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

