N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer
- PMID: 27728805
- PMCID: PMC5540451
- DOI: 10.1016/j.ccell.2016.09.005
N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer
Abstract
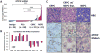
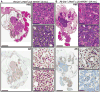
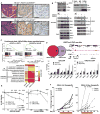
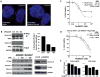
The transition from castration-resistant prostate adenocarcinoma (CRPC) to neuroendocrine prostate cancer (NEPC) has emerged as an important mechanism of treatment resistance. NEPC is associated with overexpression and gene amplification of MYCN (encoding N-Myc). N-Myc is an established oncogene in several rare pediatric tumors, but its role in prostate cancer progression is not well established. Integrating a genetically engineered mouse model and human prostate cancer transcriptome data, we show that N-Myc overexpression leads to the development of poorly differentiated, invasive prostate cancer that is molecularly similar to human NEPC. This includes an abrogation of androgen receptor signaling and induction of Polycomb Repressive Complex 2 signaling. Altogether, our data establishes N-Myc as an oncogenic driver of NEPC.
Keywords: Aurora kinase A; EZH2; N-Myc; castration-resistant prostate adenocarcinoma; genetically engineered mouse; neuroendocrine prostate cancer; prostate cancer organoid.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Neuroendocrine differentiation in prostate cancer: key epigenetic players.Transl Cancer Res. 2017 Feb;6(Suppl 1):S104-S108. doi: 10.21037/tcr.2017.01.20. Transl Cancer Res. 2017. PMID: 30613478 Free PMC article. No abstract available.
References
-
- Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

