p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer
- PMID: 27728806
- PMCID: PMC5081228
- DOI: 10.1016/j.ccell.2016.09.004
p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer
Abstract
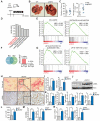
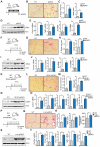
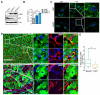
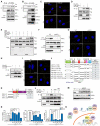
Hepatic stellate cells (HSCs) play critical roles in liver fibrosis and hepatocellular carcinoma (HCC). Vitamin D receptor (VDR) activation in HSCs inhibits liver inflammation and fibrosis. We found that p62/SQSTM1, a protein upregulated in liver parenchymal cells but downregulated in HCC-associated HSCs, negatively controls HSC activation. Total body or HSC-specific p62 ablation potentiates HSCs and enhances inflammation, fibrosis, and HCC progression. p62 directly interacts with VDR and RXR promoting their heterodimerization, which is critical for VDR:RXR target gene recruitment. Loss of p62 in HSCs impairs the repression of fibrosis and inflammation by VDR agonists. This demonstrates that p62 is a negative regulator of liver inflammation and fibrosis through its ability to promote VDR signaling in HSCs, whose activation supports HCC.
Keywords: fibrosis; hepatic stellate cells; hepatocellular carcinoma; inflammation; liver cancer; non-alcoholic steatohepatitis; nuclear receptors; p62; sequestosome-1; vitamin D receptor.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



Comment in
-
p62 in Liver Disease: Good Guy or Bad Guy?Cancer Cell. 2016 Oct 10;30(4):509-510. doi: 10.1016/j.ccell.2016.09.013. Cancer Cell. 2016. PMID: 27728799
References
-
- Anstee QM, Day CP. The genetics of NAFLD. Nature reviews Gastroenterology & hepatology. 2013;10:645–655. - PubMed
-
- Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature reviews Cancer. 2004;4:579–591. - PubMed
-
- Denk H, Stumptner C, Fuchsbichler A, Muller T, Farr G, Muller W, Terracciano L, Zatloukal K. Are the Mallory bodies and intracellular hyaline bodies in neoplastic and non-neoplastic hepatocytes related? J Pathol. 2006;208:653–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
