A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and β repertoires and identifying potential new invariant TCR α chains
- PMID: 27729009
- PMCID: PMC5059964
- DOI: 10.1186/s12865-016-0177-5
A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and β repertoires and identifying potential new invariant TCR α chains
Abstract
Background: High-throughput sequencing of T cell receptor (TCR) genes is a powerful tool for analyses of antigen specificity, clonality and diversity of T lymphocytes. Here, we developed a new TCR repertoire analysis method using 454 DNA sequencing technology in combination with an adaptor-ligation mediated polymerase chain reaction (PCR). This method allows the amplification of all TCR genes without PCR bias. To compare gene usage, diversity and similarity of expressed TCR repertoires among individuals, we conducted next-generation sequencing (NGS) of TRA and TRB genes in peripheral blood mononuclear cells from 20 healthy human individuals.
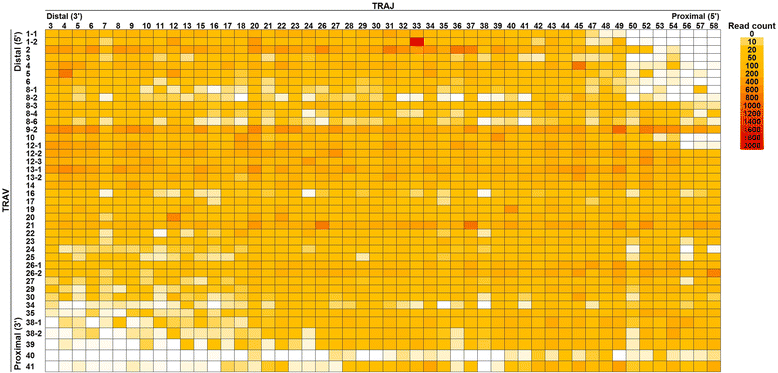
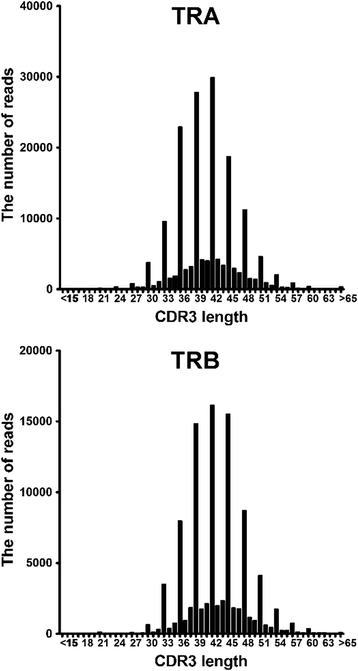
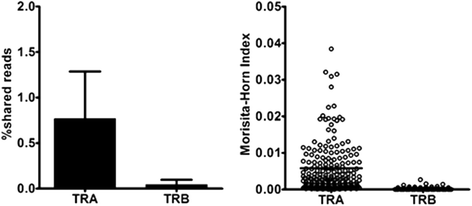
Results: From a total of 267,037 sequence reads from 20 individuals, 149,216 unique sequence reads were identified. Preferential usage of several V and J genes were observed while some recombinations of TRAV with TRAJ appeared to be restricted. The extent of TCR diversity was not significantly different between TRA and TRB, while TRA repertoires were more similar between individuals than TRB repertoires were. The interindividual similarity of TRA depended largely on the frequent presence of shared TCRs among two or more individuals. A publicly available TRA had a near-germline TCR with a shorter CDR3. Notably, shared TRA sequences, especially those shared among a large number of individuals', often contained TCRα related with invariant TCRα derived from invariant natural killer T cells and mucosal-associated invariant T cells.
Conclusion: These results suggest that retrieval of shared TCRs by NGS would be useful for the identification of potential new invariant TCRα chains. This NGS method will enable the comprehensive quantitative analysis of TCR repertoires at a clonal level.
Keywords: Immune profiling; Invariant TCRα; Next generation sequencing; Repertoire; T cell receptor.
Figures






References
-
- Tembhare P, Yuan CM, Xi L, Morris JC, Liewehr D, Venzon D, Janik JE, Raffeld M, Stetler-Stevenson M. Flow cytometric immunophenotypic assessment of T-cell clonality by Vbeta repertoire analysis: detection of T-cell clonality at diagnosis and monitoring of minimal residual disease following therapy. Am J Clin Pathol. 2011;135(6):890–900. doi: 10.1309/AJCPV2D1DDSGJDBW. - DOI - PMC - PubMed
-
- Langerak AW, van Den Beemd R, Wolvers-Tettero IL, Boor PP, van Lochem EG, Hooijkaas H, van Dongen JJ. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood. 2001;98(1):165–73. doi: 10.1182/blood.V98.1.165. - DOI - PubMed
-
- Rebai N, Pantaleo G, Demarest JF, Ciurli C, Soudeyns H, Adelsberger JW, Vaccarezza M, Walker RE, Sekaly RP, Fauci AS. Analysis of the T-cell receptor beta-chain variable-region (V beta) repertoire in monozygotic twins discordant for human immunodeficiency virus: evidence for perturbations of specific V beta segments in CD4+ T cells of the virus-positive twins. Proc Natl Acad Sci U S A. 1994;91(4):1529–33. doi: 10.1073/pnas.91.4.1529. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

