Carotenogenesis Is Regulated by 5'UTR-Mediated Translation of Phytoene Synthase Splice Variants
- PMID: 27729470
- PMCID: PMC5129717
- DOI: 10.1104/pp.16.01262
Carotenogenesis Is Regulated by 5'UTR-Mediated Translation of Phytoene Synthase Splice Variants
Abstract
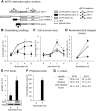
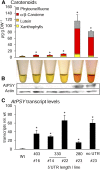
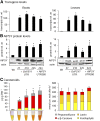
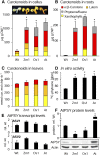
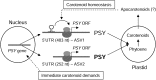
Phytoene synthase (PSY) catalyzes the highly regulated, frequently rate-limiting synthesis of the first biosynthetically formed carotene. While PSY constitutes a small gene family in most plant taxa, the Brassicaceae, including Arabidopsis (Arabidopsis thaliana), predominantly possess a single PSY gene. This monogenic situation is compensated by the differential expression of two alternative splice variants (ASV), which differ in length and in the exon/intron retention of their 5'UTRs. ASV1 contains a long 5'UTR (untranslated region) and is involved in developmentally regulated carotenoid formation, such as during deetiolation. ASV2 contains a short 5'UTR and is preferentially induced when an immediate increase in the carotenoid pathway flux is required, such as under salt stress or upon sudden light intensity changes. We show that the long 5'UTR of ASV1 is capable of attenuating the translational activity in response to high carotenoid pathway fluxes. This function resides in a defined 5'UTR stretch with two predicted interconvertible RNA conformations, as known from riboswitches, which might act as a flux sensor. The translation-inhibitory structure is absent from the short 5'UTR of ASV2 allowing to bypass translational inhibition under conditions requiring rapidly increased pathway fluxes. The mechanism is not found in the rice (Oryza sativa) PSY1 5'UTR, consistent with the prevalence of transcriptional control mechanisms in taxa with multiple PSY genes. The translational control mechanism identified is interpreted in terms of flux adjustments needed in response to retrograde signals stemming from intermediates of the plastid-localized carotenoid biosynthesis pathway.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics Int. 11: 36–42
-
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 - PubMed
-
- Alexandrov NN, Troukhan ME, Brover VV, Tatarinova T, Flavell RB, Feldmann KA (2006) Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol 60: 69–85 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

