Dosage Compensation in Drosophila-a Model for the Coordinate Regulation of Transcription
- PMID: 27729494
- PMCID: PMC5068838
- DOI: 10.1534/genetics.115.185108
Dosage Compensation in Drosophila-a Model for the Coordinate Regulation of Transcription
Abstract
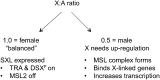
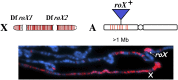
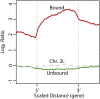
The sex chromosomes have special significance in the history of genetics. The chromosomal basis of inheritance was firmly established when Calvin Bridges demonstrated that exceptions to Mendel's laws of segregation were accompanied at the cytological level by exceptional sex chromosome segregation. The morphological differences between X and Y exploited in Bridges' experiments arose as a consequence of the evolution of the sex chromosomes. Originally a homologous chromosome pair, the degeneration of the Y chromosome has been accompanied by a requirement for increased expression of the single X chromosome in males. Drosophila has been a model for the study of this dosage compensation and has brought key strengths, including classical genetics, the exceptional cytology of polytene chromosomes, and more recently, comprehensive genomics. The impact of these studies goes beyond sex chromosome regulation, providing valuable insights into mechanisms for the establishment and maintenance of chromatin domains, and for the coordinate regulation of transcription.
Keywords: Drosophila; FlyBook; X chromosome; dosage dependence; genetic screens; male-specific lethal.
Copyright © 2016 by the Genetics Society of America.
Figures





References
-
- Akhtar A., Gasser S. M., 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8: 507–517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

