Evolution of codon usage in Zika virus genomes is host and vector specific
- PMID: 27729643
- PMCID: PMC5117728
- DOI: 10.1038/emi.2016.106
Evolution of codon usage in Zika virus genomes is host and vector specific
Abstract
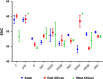
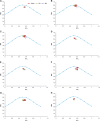
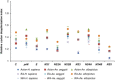
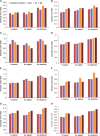
The codon usage patterns of viruses reflect the evolutionary changes that allow them to optimize their survival and adapt their fitness to the external environment and, most importantly, their hosts. Here we report the genotype-specific codon usage patterns of Zika virus (ZIKV) strains from the current and previous outbreaks. Several genotype-specific and common codon usage traits were noted in the ZIKV coding sequences, indicating their independent evolutionary origins from a common ancestor. The overall influence of natural selection was more profound than that of mutation pressure, acting on a specific set of viral genes in the Asian-genotype ZIKV strains from the recent outbreak. An interplay between codon adaptation and deoptimization may have allowed the virus to adapt to multiple host and vectors and is reported for the first time in ZIKV genomes. Combining our codon analysis with geographical data on Aedes populations in the Americas suggested that ZIKV has evolved host- and vector-specific codon usage patterns to maintain successful replication and transmission chains within multiple hosts and vectors.
Figures





References
-
- Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol 2007; 152: 687–696. - PubMed
-
- Duffy MR, Chen TH, Hancock WT, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 2009; 360: 2536–2543. - PubMed
-
- Cao-Lormeau VM, Musso D. Emerging arboviruses in the Pacific. Lancet 2014; 384: 1571–1572. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
