Using FlyBase, a Database of Drosophila Genes and Genomes
- PMID: 27730573
- PMCID: PMC5107610
- DOI: 10.1007/978-1-4939-6371-3_1
Using FlyBase, a Database of Drosophila Genes and Genomes
Abstract
For nearly 25 years, FlyBase (flybase.org) has provided a freely available online database of biological information about Drosophila species, focusing on the model organism D. melanogaster. The need for a centralized, integrated view of Drosophila research has never been greater as advances in genomic, proteomic, and high-throughput technologies add to the quantity and diversity of available data and resources.FlyBase has taken several approaches to respond to these changes in the research landscape. Novel report pages have been generated for new reagent types and physical interaction data; Drosophila models of human disease are now represented and showcased in dedicated Human Disease Model Reports; other integrated reports have been established that bring together related genes, datasets, or reagents; Gene Reports have been revised to improve access to new data types and to highlight functional data; links to external sites have been organized and expanded; and new tools have been developed to display and interrogate all these data, including improved batch processing and bulk file availability. In addition, several new community initiatives have served to enhance interactions between researchers and FlyBase, resulting in direct user contributions and improved feedback.This chapter provides an overview of the data content, organization, and available tools within FlyBase, focusing on recent improvements. We hope it serves as a guide for our diverse user base, enabling efficient and effective exploration of the database and thereby accelerating research discoveries.
Keywords: Database; Drosophila; FlyBase; Genetics; Genomics; Translational research.
Figures

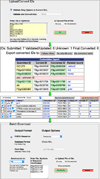
References
-
- Drysdale R FlyBase Consortium. FlyBase : a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

