No Association of Coronary Artery Disease with X-Chromosomal Variants in Comprehensive International Meta-Analysis
- PMID: 27731410
- PMCID: PMC5059659
- DOI: 10.1038/srep35278
No Association of Coronary Artery Disease with X-Chromosomal Variants in Comprehensive International Meta-Analysis
Abstract
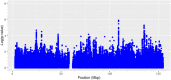
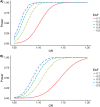
In recent years, genome-wide association studies have identified 58 independent risk loci for coronary artery disease (CAD) on the autosome. However, due to the sex-specific data structure of the X chromosome, it has been excluded from most of these analyses. While females have 2 copies of chromosome X, males have only one. Also, one of the female X chromosomes may be inactivated. Therefore, special test statistics and quality control procedures are required. Thus, little is known about the role of X-chromosomal variants in CAD. To fill this gap, we conducted a comprehensive X-chromosome-wide meta-analysis including more than 43,000 CAD cases and 58,000 controls from 35 international study cohorts. For quality control, sex-specific filters were used to adequately take the special structure of X-chromosomal data into account. For single study analyses, several logistic regression models were calculated allowing for inactivation of one female X-chromosome, adjusting for sex and investigating interactions between sex and genetic variants. Then, meta-analyses including all 35 studies were conducted using random effects models. None of the investigated models revealed genome-wide significant associations for any variant. Although we analyzed the largest-to-date sample, currently available methods were not able to detect any associations of X-chromosomal variants with CAD.
Figures
References
-
- König I. R., Loley C., Erdmann J. & Ziegler A. How to include chromosome X in your genome-wide association study. Genet. Epidemiol. 38, 97–103 (2014). - PubMed
-
- Devlin B. & Roeder K. Genomic control for association studies. Biometrics 55, 997–1004 (1999). - PubMed
-
- Hindorff L. et al. A Catalog of Published Genome-Wide Association Studies www.genome.gov/gwastudies (2014).
Publication types
MeSH terms
Grants and funding
- U01 HG007417/HG/NHGRI NIH HHS/United States
- R01 HL087676/HL/NHLBI NIH HHS/United States
- G0700931/MRC_/Medical Research Council/United Kingdom
- R01 DK082766/DK/NIDDK NIH HHS/United States
- MOP-2380941 /CIHR/Canada
- MOP82810/CIHR/Canada
- 084723/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL087647/HL/NHLBI NIH HHS/United States
- R01 HL107816/HL/NHLBI NIH HHS/United States
- R01 HL095987/HL/NHLBI NIH HHS/United States
- P01 HL076491/HL/NHLBI NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- G0601966/MRC_/Medical Research Council/United Kingdom
- MOP77682/CIHR/Canada
- K23 DK088942/DK/NIDDK NIH HHS/United States
- RC2 HL101621/HL/NHLBI NIH HHS/United States
- 07611/WT_/Wellcome Trust/United Kingdom
- RG/14/5/30893/BHF_/British Heart Foundation/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

